4. Data Exploration¶
Note
Know yourself and know your enemy, and you will never be defeated – idiom, from Sunzi’s Art of War
4.1. Procedures¶
Data mining is a complex process that aims to discover patterns in large data sets starting from a collection of exsting data. In my opinion, data minig contains four main steps:
- Collecting data: This is a complex step, I will assume we have already gotten the datasets.
- Pre-processing: In this step, we need to try to understand your data, denoise, do dimentation reduction and select proper predictors etc.
- Feeding data mining: In this step, we need to use your data to feed your model.
- Post-processing : In this step, we need to interpret and evaluate your model.
In this section, we will try to know our enemy – datasets. We will learn how to load data, how to understand data with statistics method and how to underdtand data with visualization. Next, we will start with Loading Datasets for the Pre-processing.
4.2. Datasets in this Tutorial¶
The datasets for this tutorial are available to download: Heart, Energy Efficienency. Those data are from my course matrials, the copyrights blongs to the origial authors.
4.3. Loading Datasets¶
There are three main data source database, *.csv and *.xlsx. We will show how to load those two types of data in R and Python, respectively.
4.3.1. Loading table format database¶
User and Database information:
user = '*******'
pw='********'
host = '**.***.***.**'
database = '**'
table_name = '***'
Python
- Loading data from database in Python
# import library
import psycopg2
import pandas as pd
# Create the database connection
conn = psycopg2.connect(host=host, database=database,
user=user, password=pw)
cur = conn.cursor()
# Create the SQL query string.
sql = """
SELECT *
FROM {table_name}
""".format(table_name=table_name)
df = pd.read_sql(sql, conn)
df.head(4)
R
- Loading data from database in R
# load the library
library("sqldf")
library('RODBC')
library('RPostgreSQL')
# Create a driver
drv <- DBI::dbDriver( "PostgreSQL" )
# Create the database connection
conn <- dbConnect( drv, dbname = database, host = host,port = '5432',
user = user, password = pw )
# Create the SQL query string. Include a semi-colon to terminate
querystring = sprintf('SELECT * FROM %s;', table_name)
# Execute the query and return results as a data frame
df = dbGetQuery(conn, querystring )
head(df)
4.3.2. Loading data from .csv¶
Python
- Loading data from
.csvin Python
import pandas as pd
# set data path
path ='~/Dropbox/MachineLearningAlgorithms/python_code/data/Heart.csv'
# read data set
rawdata = pd.read_csv(path)
R
- Loading data from
.csvin R
# set the path or enverionment
setwd("/home/feng/R-language/sat577/HW#4/data")
# read data set
rawdata = read.csv("spam.csv")
4.3.3. Loading data from .xlsx¶
Python
- Loading data from
.xlsxin Python
import pandas as pd
# set data path
path = ('/home/feng/Dropbox/MachineLearningAlgorithms/python_code/data/'
'energy_efficiency.xlsx')
# read data set from first sheet
rawdata= pd.read_excel(path,sheetname=0)
R
- Loading data from
.xlsxin R
# set the path or enverionment
setwd("~/Dropbox/R-language/sat577/")
#install.packages("readxl") # CRAN version
library(readxl)
# read data set
energy_eff=read_excel("energy_efficiency.xlsx")
4.4. Audit Data¶
In my opinion, data auditing is the first step you need to do when you get your dataset. Since you need to know whether the data quality is good enough or not. My PyAudit: Python Data Audit Library can be found at: PyAudit. You can install the PyAudit from [PyPI](https://pypi.org/project/PyAudit):
pip install PyAudit
4.4.1. Check missing rate¶
Python
- Checking missing rate in Python
import pandas as pd
d = {'A': [1, 2, None, 3],
'B': [None, None, 4, 5],
'C': [None, 'b', 'c', 'd']}
# create DataFrame
df = pd.DataFrame(d)
print(df)
# define the missing rate function
def missing_rate(df_in):
# calculate missing rate and transpose the DataFrame
rate = df_in.isnull().sum() / df_in.shape[0]
# rename the column
rate = pd.DataFrame(rate).reset_index()\
.rename(columns={'index': 'feature', 0: 'missing_rate'})
print(rate)
missing_rate(df)
The results:
A B C
0 1.0 NaN None
1 2.0 NaN b
2 NaN 4.0 c
3 3.0 5.0 d
feature missing_rate
0 A 0.25
1 B 0.50
2 C 0.25
R
- Checking missing rate in R
# create DataFrame
x = data.frame(A = c(1, 2, NA, 3), B = c(NA, NA, 4, 5), C = c(NA, 'b', 'c', 'd'))
# loding library
library('dplyr')
#library('tidyverse')
# define the missing rate function
missing_rate <- function(df){
# calculate missing rate and transpose the DataFrame
rate <-t( df %>% summarize_all(funs(sum(is.na(.)) / length(.))))
# rename the column
colnames(rate)[1] <- "missing_rate"
print(rate)
}
x
missing_rate(x)
The results:
> x
A B C
1 1 NA <NA>
2 2 NA b
3 NA 4 c
4 3 5 d
> missing_rate(x)
missing_rate
A 0.25
B 0.50
C 0.25
4.4.2. Checking zero variance features¶
Python
- Checking zero variance features in Python
import pandas as pd
d = {'A': [1, 2, 3, 3],
'B': [1, 1, 1, 1],
'C': ['a', 'b', 'c', 'd']}
# create DataFrame
df = pd.DataFrame(d)
print(df)
def zero_variance(df_in):
counts = df_in.nunique()
counts = pd.DataFrame(counts)\
.reset_index().rename(columns={'index': 'feature', 0: 'count'})
return list(counts[counts['count'] == 1]['feature'])
print(zero_variance(df))
A B C
0 1 1 a
1 2 1 b
2 3 1 c
3 3 1 d
['B']
R
- Checking zero variance features in R
df = data.frame(A = c(1, 2, 3, 3), B = c(1, 1, 1, 1), C = c('a', 'b', 'c', 'd'))
zero_variance <- function(df){
compData <- data.frame(feature= c(NA), count= c(NA))
for(i in 1:ncol(df))
{
compData[i, ] <- c(colnames(df)[i],length(unique(df[,i])))
}
return(compData[compData$count==1,]$feature)
}
> zero_variance(df)
[1] "B"
4.5. Understand Data With Statistics methods¶
After we get the data in hand, then we can try to understand them. I will use “Heart.csv” dataset as a example to demonstrate how to use those statistics methods.
4.5.1. Summary of the data¶
It is always good to have a glance over the summary of the data. Since from the summary you will know some statistics features of your data, and you will also know whether you data contains missing data or not.
Python
- Summary of the data in Python
print("> data summary")
print rawdata.describe()
Then you will get
> data summary
Age Sex RestBP Chol Fbs RestECG \
count 303.000000 303.000000 303.000000 303.000000 303.000000 303.000000
mean 54.438944 0.679868 131.689769 246.693069 0.148515 0.990099
std 9.038662 0.467299 17.599748 51.776918 0.356198 0.994971
min 29.000000 0.000000 94.000000 126.000000 0.000000 0.000000
25% 48.000000 0.000000 120.000000 211.000000 0.000000 0.000000
50% 56.000000 1.000000 130.000000 241.000000 0.000000 1.000000
75% 61.000000 1.000000 140.000000 275.000000 0.000000 2.000000
max 77.000000 1.000000 200.000000 564.000000 1.000000 2.000000
MaxHR ExAng Oldpeak Slope Ca
count 303.000000 303.000000 303.000000 303.000000 299.000000
mean 149.607261 0.326733 1.039604 1.600660 0.672241
std 22.875003 0.469794 1.161075 0.616226 0.937438
min 71.000000 0.000000 0.000000 1.000000 0.000000
25% 133.500000 0.000000 0.000000 1.000000 0.000000
50% 153.000000 0.000000 0.800000 2.000000 0.000000
75% 166.000000 1.000000 1.600000 2.000000 1.000000
max 202.000000 1.000000 6.200000 3.000000 3.000000
R
- Summary of the data in R
summary(rawdata)
Then you will get
> summary(rawdata)
Age Sex ChestPain RestBP
Min. :29.00 Min. :0.0000 asymptomatic:144 Min. : 94.0
1st Qu.:48.00 1st Qu.:0.0000 nonanginal : 86 1st Qu.:120.0
Median :56.00 Median :1.0000 nontypical : 50 Median :130.0
Mean :54.44 Mean :0.6799 typical : 23 Mean :131.7
3rd Qu.:61.00 3rd Qu.:1.0000 3rd Qu.:140.0
Max. :77.00 Max. :1.0000 Max. :200.0
Chol Fbs RestECG MaxHR
Min. :126.0 Min. :0.0000 Min. :0.0000 Min. : 71.0
1st Qu.:211.0 1st Qu.:0.0000 1st Qu.:0.0000 1st Qu.:133.5
Median :241.0 Median :0.0000 Median :1.0000 Median :153.0
Mean :246.7 Mean :0.1485 Mean :0.9901 Mean :149.6
3rd Qu.:275.0 3rd Qu.:0.0000 3rd Qu.:2.0000 3rd Qu.:166.0
Max. :564.0 Max. :1.0000 Max. :2.0000 Max. :202.0
ExAng Oldpeak Slope Ca
Min. :0.0000 Min. :0.00 Min. :1.000 Min. :0.0000
1st Qu.:0.0000 1st Qu.:0.00 1st Qu.:1.000 1st Qu.:0.0000
Median :0.0000 Median :0.80 Median :2.000 Median :0.0000
Mean :0.3267 Mean :1.04 Mean :1.601 Mean :0.6722
3rd Qu.:1.0000 3rd Qu.:1.60 3rd Qu.:2.000 3rd Qu.:1.0000
Max. :1.0000 Max. :6.20 Max. :3.000 Max. :3.0000
NA's :4
Thal AHD
fixed : 18 No :164
normal :166 Yes:139
reversable:117
NA's : 2
4.5.2. The size of the data¶
Most of time, we also need to know the size or dimension of our data. Such as when you need to extract the response from the dataset, you need the number of column, or when you try to split your data into train and test data set, you need know the number of row.
Python
- Checking size in Python
nrow, ncol = rawdata.shape
print nrow, ncol
or you can use the follwing code
nrow=rawdata.shape[0] #gives number of row count
ncol=rawdata.shape[1] #gives number of col count
print(nrow, ncol)
Then you will get
Raw data size
303 14
R
- Checking size in R
dim(rawdata)
Or you can use the following code
nrow=nrow(rawdata)
ncol=ncol(rawdata)
c(nrow, ncol)
Then you will get
> dim(rawdata)
[1] 303 14
4.5.3. Data type of the features¶
Data type is also very important, since some functions or methods can not be applied to the qualitative data or some machine learning algorithm will take some types as categorical data, you need to remove those features or transform them into quantitative data.
Python
- Checking data type in Pyhton
print(rawdata.dtypes)
Then you will get
Data Format:
Age int64
Sex int64
ChestPain object
RestBP int64
Chol int64
Fbs int64
RestECG int64
MaxHR int64
ExAng int64
Oldpeak float64
Slope int64
Ca float64
Thal object
AHD object
dtype: object
R
- Checking data format in R
# install the package
install.packages("mlbench")
library(mlbench)
sapply(rawdata, class)
Then you will get
> sapply(rawdata, class)
Age Sex ChestPain RestBP Chol Fbs RestECG
"integer" "integer" "factor" "integer" "integer" "integer" "integer"
MaxHR ExAng Oldpeak Slope Ca Thal AHD
"integer" "integer" "numeric" "integer" "integer" "factor" "factor"
4.5.4. The column names¶
Python
- Checking column names of the data in Python
colNames = rawdata.columns.tolist()
print "Column names:"
print colNames
Then you will get
Column names:
['Age', 'Sex', 'ChestPain', 'RestBP', 'Chol', 'Fbs', 'RestECG', 'MaxHR',
'ExAng', 'Oldpeak', 'Slope', 'Ca', 'Thal', 'AHD']
R
- Checking column names of the data in R
colnames(rawdata)
attach(rawdata) # enable you can directly use name as features
Then you will get
> colnames(rawdata)
[1] "Age" "Sex" "ChestPain" "RestBP" "Chol"
[6] "Fbs" "RestECG" "MaxHR" "ExAng" "Oldpeak"
[11] "Slope" "Ca" "Thal" "AHD"
4.5.5. The first or last parts of the data¶
Python
- Checking first parts of the data in Python
print("\n Sample data:")
print(rawdata.head(6))
Then you will get
Sample data:
Age Sex ChestPain RestBP Chol Fbs RestECG MaxHR ExAng Oldpeak \
0 63 1 typical 145 233 1 2 150 0 2.3
1 67 1 asymptomatic 160 286 0 2 108 1 1.5
2 67 1 asymptomatic 120 229 0 2 129 1 2.6
3 37 1 nonanginal 130 250 0 0 187 0 3.5
4 41 0 nontypical 130 204 0 2 172 0 1.4
5 56 1 nontypical 120 236 0 0 178 0 0.8
Slope Ca Thal AHD
0 3 0 fixed No
1 2 3 normal Yes
2 2 2 reversable Yes
3 3 0 normal No
4 1 0 normal No
5 1 0 normal No
R
- Checking first parts of the data in R
head(rawdata)
Then you will get
> head(rawdata)
Age Sex ChestPain RestBP Chol Fbs RestECG MaxHR ExAng Oldpeak
1 63 1 typical 145 233 1 2 150 0 2.3
2 67 1 asymptomatic 160 286 0 2 108 1 1.5
3 67 1 asymptomatic 120 229 0 2 129 1 2.6
4 37 1 nonanginal 130 250 0 0 187 0 3.5
5 41 0 nontypical 130 204 0 2 172 0 1.4
6 56 1 nontypical 120 236 0 0 178 0 0.8
Slope Ca Thal AHD
1 3 0 fixed No
2 2 3 normal Yes
3 2 2 reversable Yes
4 3 0 normal No
5 1 0 normal No
6 1 0 normal No
You can use the samilar way (tail) to check the last part of the data, for simplicity, i will skip it.
4.5.6. Correlation Matrix¶
Python
- Computing correlation matrix in Python
print("\n correlation Matrix")
print(rawdata.corr())
Then you will get
correlation Matrix
Age Sex RestBP Chol Fbs RestECG MaxHR \
Age 1.000000 -0.097542 0.284946 0.208950 0.118530 0.148868 -0.393806
Sex -0.097542 1.000000 -0.064456 -0.199915 0.047862 0.021647 -0.048663
RestBP 0.284946 -0.064456 1.000000 0.130120 0.175340 0.146560 -0.045351
Chol 0.208950 -0.199915 0.130120 1.000000 0.009841 0.171043 -0.003432
Fbs 0.118530 0.047862 0.175340 0.009841 1.000000 0.069564 -0.007854
RestECG 0.148868 0.021647 0.146560 0.171043 0.069564 1.000000 -0.083389
MaxHR -0.393806 -0.048663 -0.045351 -0.003432 -0.007854 -0.083389 1.000000
ExAng 0.091661 0.146201 0.064762 0.061310 0.025665 0.084867 -0.378103
Oldpeak 0.203805 0.102173 0.189171 0.046564 0.005747 0.114133 -0.343085
Slope 0.161770 0.037533 0.117382 -0.004062 0.059894 0.133946 -0.385601
Ca 0.362605 0.093185 0.098773 0.119000 0.145478 0.128343 -0.264246
ExAng Oldpeak Slope Ca
Age 0.091661 0.203805 0.161770 0.362605
Sex 0.146201 0.102173 0.037533 0.093185
RestBP 0.064762 0.189171 0.117382 0.098773
Chol 0.061310 0.046564 -0.004062 0.119000
Fbs 0.025665 0.005747 0.059894 0.145478
RestECG 0.084867 0.114133 0.133946 0.128343
MaxHR -0.378103 -0.343085 -0.385601 -0.264246
ExAng 1.000000 0.288223 0.257748 0.145570
Oldpeak 0.288223 1.000000 0.577537 0.295832
Slope 0.257748 0.577537 1.000000 0.110119
Ca 0.145570 0.295832 0.110119 1.000000
R
- Computing correlation matrix in R
# get numerical data and remove NAN
numdata=na.omit(rawdata[,c(1:2,4:12)])
# computing correlation matrix
cor(numdata)
Then you will get
> cor(numdata)
Age Sex RestBP Chol Fbs
Age 1.00000000 -0.09181347 0.29069633 0.203376601 0.128675921
Sex -0.09181347 1.00000000 -0.06552127 -0.195907357 0.045861783
RestBP 0.29069633 -0.06552127 1.00000000 0.132284171 0.177623291
Chol 0.20337660 -0.19590736 0.13228417 1.000000000 0.006664176
Fbs 0.12867592 0.04586178 0.17762329 0.006664176 1.000000000
RestECG 0.14974915 0.02643577 0.14870922 0.164957542 0.058425836
MaxHR -0.39234176 -0.05206445 -0.04805281 0.002179081 -0.003386615
ExAng 0.09510850 0.14903849 0.06588463 0.056387955 0.011636935
Oldpeak 0.19737552 0.11023676 0.19161540 0.040430535 0.009092935
Slope 0.15895990 0.03933739 0.12110773 -0.009008239 0.053776677
Ca 0.36260453 0.09318476 0.09877326 0.119000487 0.145477522
RestECG MaxHR ExAng Oldpeak Slope
Age 0.14974915 -0.392341763 0.09510850 0.197375523 0.158959901
Sex 0.02643577 -0.052064447 0.14903849 0.110236756 0.039337394
RestBP 0.14870922 -0.048052805 0.06588463 0.191615405 0.121107727
Chol 0.16495754 0.002179081 0.05638795 0.040430535 -0.009008239
Fbs 0.05842584 -0.003386615 0.01163693 0.009092935 0.053776677
RestECG 1.00000000 -0.077798148 0.07408360 0.110275054 0.128907169
MaxHR -0.07779815 1.000000000 -0.37635897 -0.341262236 -0.381348495
ExAng 0.07408360 -0.376358975 1.00000000 0.289573103 0.254302081
Oldpeak 0.11027505 -0.341262236 0.28957310 1.000000000 0.579775260
Slope 0.12890717 -0.381348495 0.25430208 0.579775260 1.000000000
Ca 0.12834265 -0.264246253 0.14556960 0.295832115 0.110119188
Ca
Age 0.36260453
Sex 0.09318476
RestBP 0.09877326
Chol 0.11900049
Fbs 0.14547752
RestECG 0.12834265
MaxHR -0.26424625
ExAng 0.14556960
Oldpeak 0.29583211
Slope 0.11011919
Ca 1.00000000
4.5.7. Covariance Matrix¶
Python
- Computing covariance matrix in Python
print("\n covariance Matrix")
print(rawdata.corr())
Then you will get
covariance Matrix
Age Sex RestBP Chol Fbs RestECG \
Age 81.697419 -0.411995 45.328678 97.787489 0.381614 1.338797
Sex -0.411995 0.218368 -0.530107 -4.836994 0.007967 0.010065
RestBP 45.328678 -0.530107 309.751120 118.573339 1.099207 2.566455
Chol 97.787489 -4.836994 118.573339 2680.849190 0.181496 8.811521
Fbs 0.381614 0.007967 1.099207 0.181496 0.126877 0.024654
RestECG 1.338797 0.010065 2.566455 8.811521 0.024654 0.989968
MaxHR -81.423065 -0.520184 -18.258005 -4.064651 -0.063996 -1.897941
ExAng 0.389220 0.032096 0.535473 1.491345 0.004295 0.039670
Oldpeak 2.138850 0.055436 3.865638 2.799282 0.002377 0.131850
Slope 0.901034 0.010808 1.273053 -0.129598 0.013147 0.082126
Ca 3.066396 0.040964 1.639436 5.791385 0.048394 0.119706
MaxHR ExAng Oldpeak Slope Ca
Age -81.423065 0.389220 2.138850 0.901034 3.066396
Sex -0.520184 0.032096 0.055436 0.010808 0.040964
RestBP -18.258005 0.535473 3.865638 1.273053 1.639436
Chol -4.064651 1.491345 2.799282 -0.129598 5.791385
Fbs -0.063996 0.004295 0.002377 0.013147 0.048394
RestECG -1.897941 0.039670 0.131850 0.082126 0.119706
MaxHR 523.265775 -4.063307 -9.112209 -5.435501 -5.686270
ExAng -4.063307 0.220707 0.157216 0.074618 0.064162
Oldpeak -9.112209 0.157216 1.348095 0.413219 0.322753
Slope -5.435501 0.074618 0.413219 0.379735 0.063747
Ca -5.686270 0.064162 0.322753 0.063747 0.878791
R
- Computing covariance matrix in R
# get numerical data and remove NAN
numdata=na.omit(rawdata[,c(1:2,4:12)])
# computing covariance matrix
cov(numdata)
Then you will get
> cov(numdata)
Age Sex RestBP Chol Fbs
Age 81.3775448 -0.388397567 46.4305852 95.2454603 0.411909946
Sex -0.3883976 0.219905277 -0.5440170 -4.7693542 0.007631703
RestBP 46.4305852 -0.544016969 313.4906736 121.5937353 1.116001885
Chol 95.2454603 -4.769354223 121.5937353 2695.1442616 0.122769410
Fbs 0.4119099 0.007631703 1.1160019 0.1227694 0.125923099
RestECG 1.3440551 0.012334179 2.6196943 8.5204709 0.020628044
MaxHR -81.2442706 -0.560447577 -19.5302126 2.5968104 -0.027586362
ExAng 0.4034028 0.032861215 0.5484838 1.3764001 0.001941595
Oldpeak 2.0721791 0.060162510 3.9484299 2.4427678 0.003755247
Slope 0.8855132 0.011391439 1.3241566 -0.2887926 0.011784247
Ca 3.0663958 0.040964288 1.6394357 5.7913852 0.048393975
RestECG MaxHR ExAng Oldpeak Slope
Age 1.34405513 -81.24427061 0.403402842 2.072179076 0.88551323
Sex 0.01233418 -0.56044758 0.032861215 0.060162510 0.01139144
RestBP 2.61969428 -19.53021257 0.548483760 3.948429889 1.32415658
Chol 8.52047092 2.59681040 1.376400081 2.442767839 -0.28879262
Fbs 0.02062804 -0.02758636 0.001941595 0.003755247 0.01178425
RestECG 0.98992166 -1.77682880 0.034656910 0.127690736 0.07920136
MaxHR -1.77682880 526.92866602 -4.062052479 -9.116871675 -5.40571480
ExAng 0.03465691 -4.06205248 0.221072479 0.158455478 0.07383673
Oldpeak 0.12769074 -9.11687168 0.158455478 1.354451303 0.41667415
Slope 0.07920136 -5.40571480 0.073836726 0.416674149 0.38133824
Ca 0.11970551 -5.68626967 0.064162421 0.322752576 0.06374717
Ca
Age 3.06639582
Sex 0.04096429
RestBP 1.63943570
Chol 5.79138515
Fbs 0.04839398
RestECG 0.11970551
MaxHR -5.68626967
ExAng 0.06416242
Oldpeak 0.32275258
Slope 0.06374717
Ca 0.87879060
4.6. Understand Data With Visualization¶
A picture is worth a thousand words. You will see the powerful impact of the figures in this section.
4.6.1. Summary plot of data in figure¶
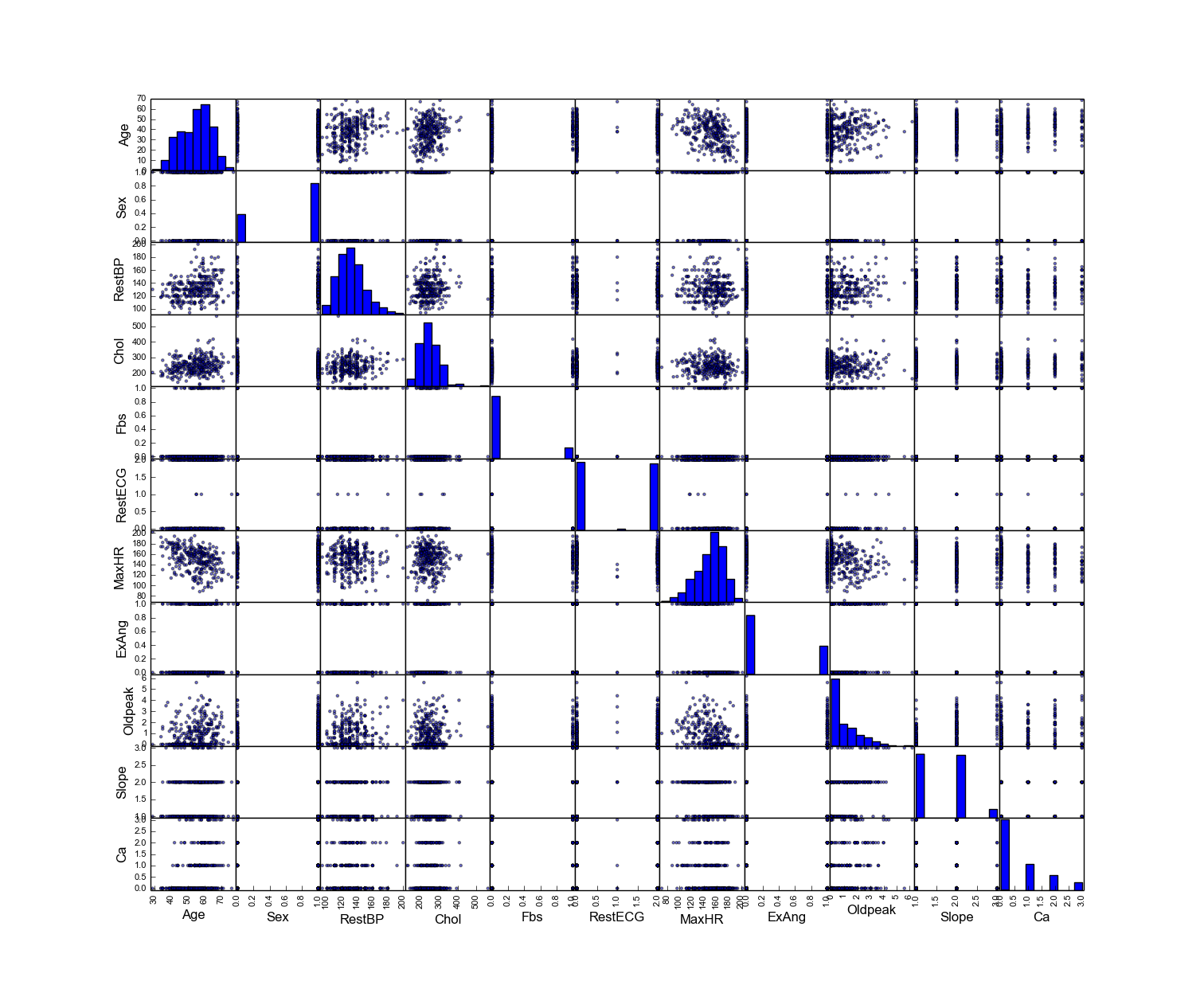
Python
- Summary plot in Python
# plot of the summary
plot(rawdata)
Then you will get Figure Summary plot of the data with Python.
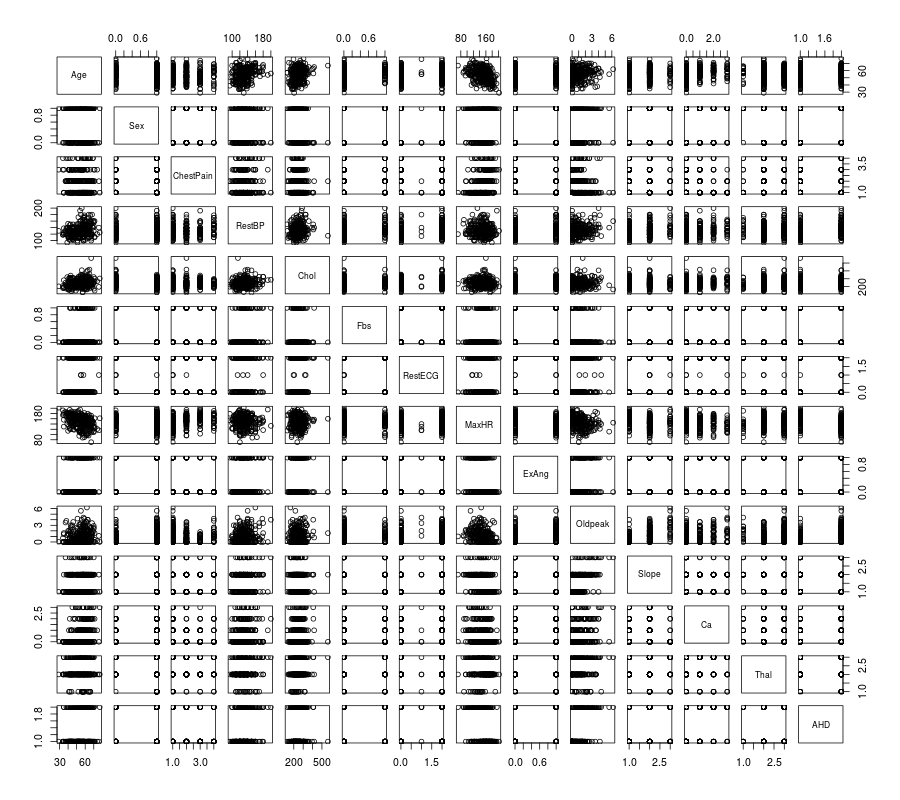
R
- Summary plot in R
# plot of the summary
plot(rawdata)
Then you will get Figure Summary plot of the data with R.
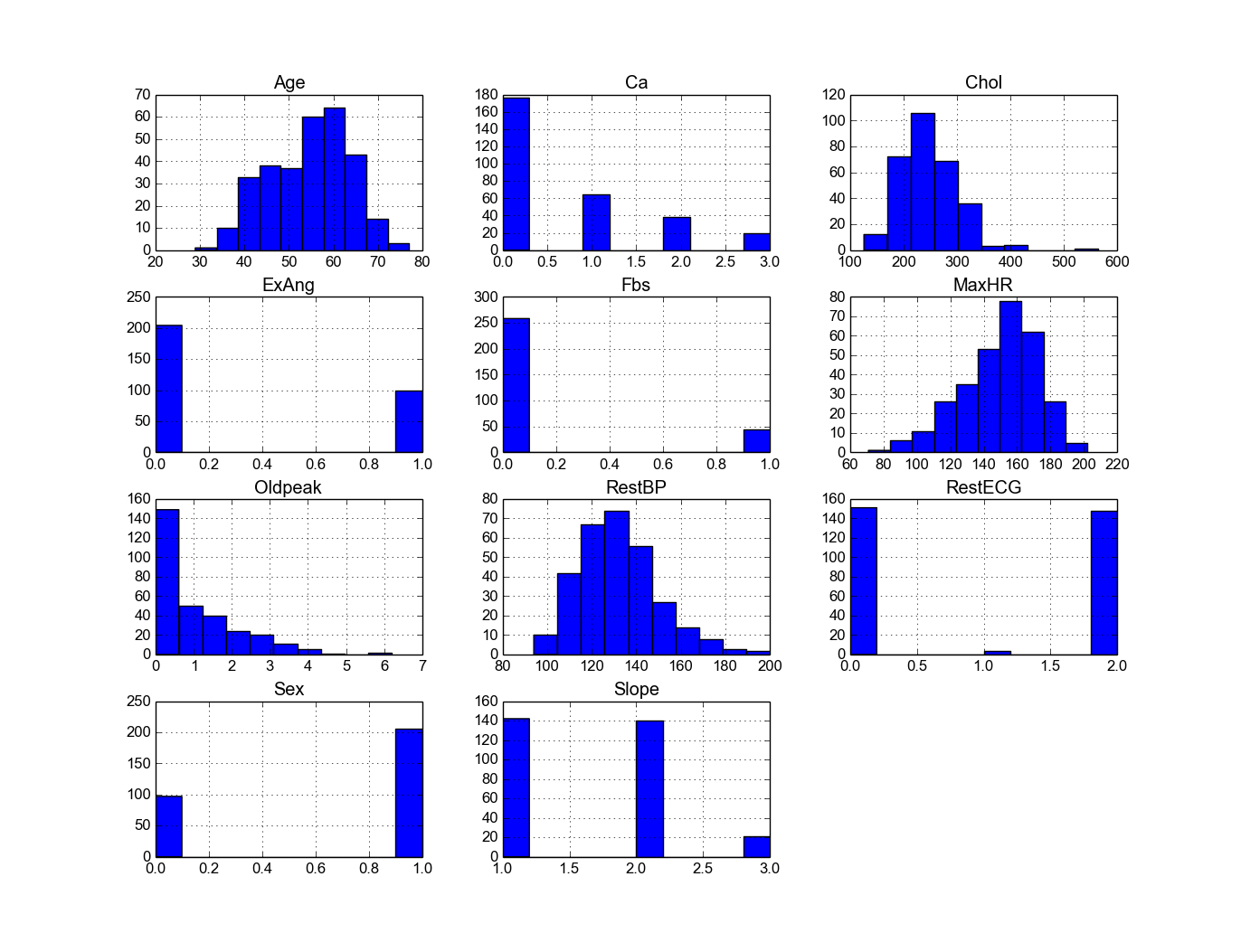
4.6.2. Histogram of the quantitative predictors¶
Python
- Histogram in Python
# Histogram
rawdata.hist()
plt.show()
Then you will get Figure Histogram in Python.
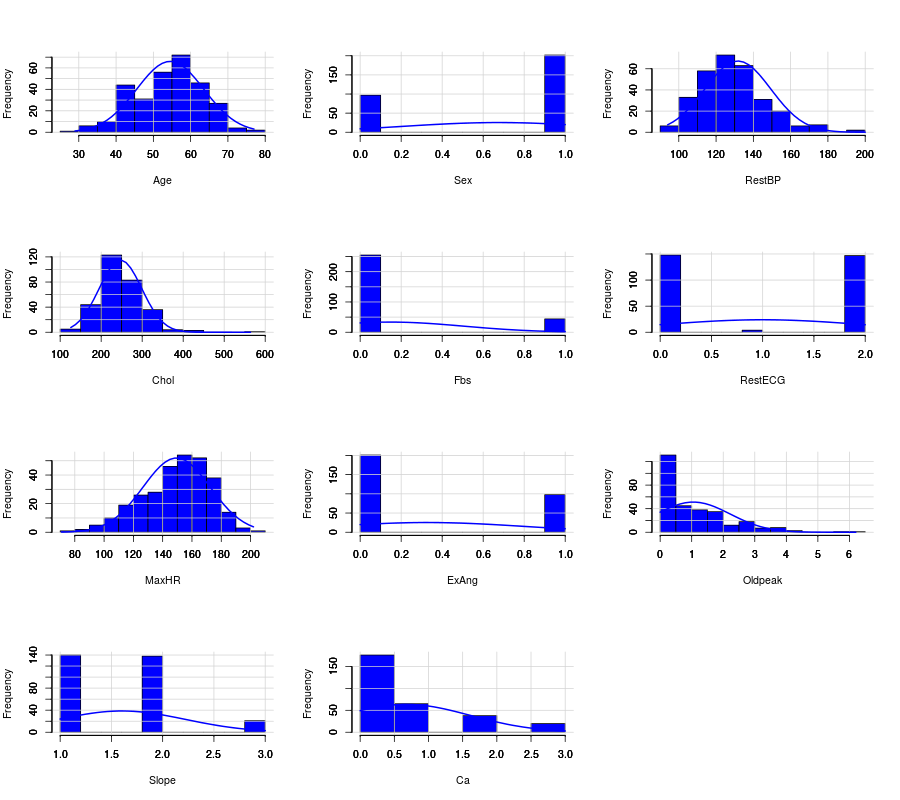
R
- Histogram in R
# Histogram with normal curve plot
dev.off()
Nvars=ncol(numdata)
name=colnames(numdata)
par(mfrow =c (4,3))
for (i in 1:Nvars)
{
x<- numdata[,i]
h<-hist(x, breaks=10, freq=TRUE, col="blue", xlab=name[i],main=" ",
font.lab=1)
axis(1, tck=1, col.ticks="light gray")
axis(1, tck=-0.015, col.ticks="black")
axis(2, tck=1, col.ticks="light gray", lwd.ticks="1")
axis(2, tck=-0.015)
xfit<-seq(min(x),max(x),length=40)
yfit<-dnorm(xfit,mean=mean(x),sd=sd(x))
yfit <- yfit*diff(h$mids[1:2])*length(x)
lines(xfit, yfit, col="blue", lwd=2)
}
Then you will get Figure Histogram with normal curve plot in R.
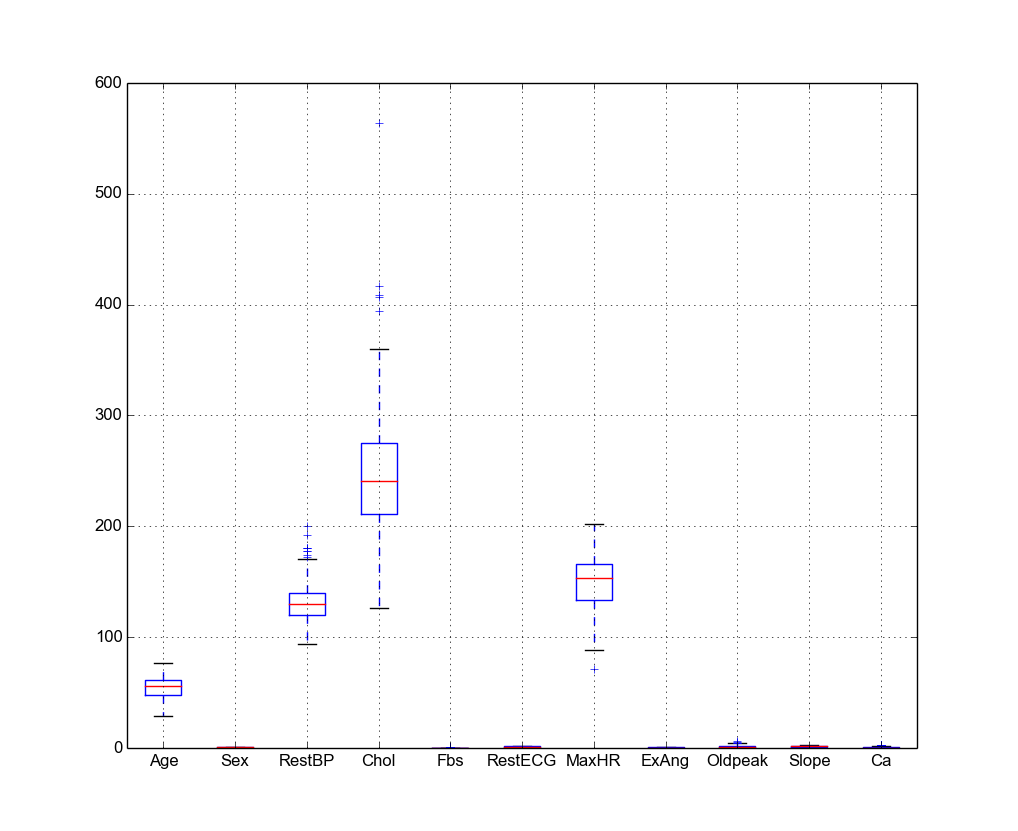
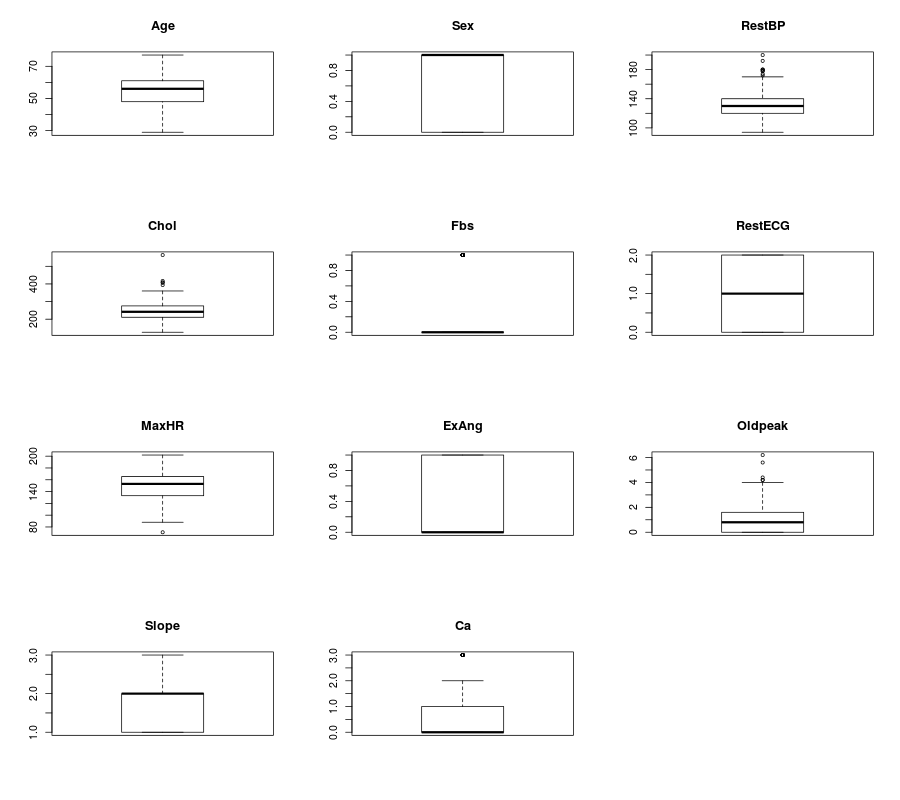
4.6.3. Boxplot of the quantitative predictors¶
Python
- Boxplot in Python
# boxplot
pd.DataFrame.boxplot(rawdata)
plt.show()
Then you will get Figure Histogram in Python.
R
- Boxplot in R
dev.off()
name=colnames(numdata)
Nvars=ncol(numdata)
# boxplot
par(mfrow =c (4,3))
for (i in 1:Nvars)
{
#boxplot(numdata[,i]~numdata[,Nvars],data=data,main=name[i])
boxplot(numdata[,i],data=numdata,main=name[i])
}
Then you will get Figure Boxplots in R.
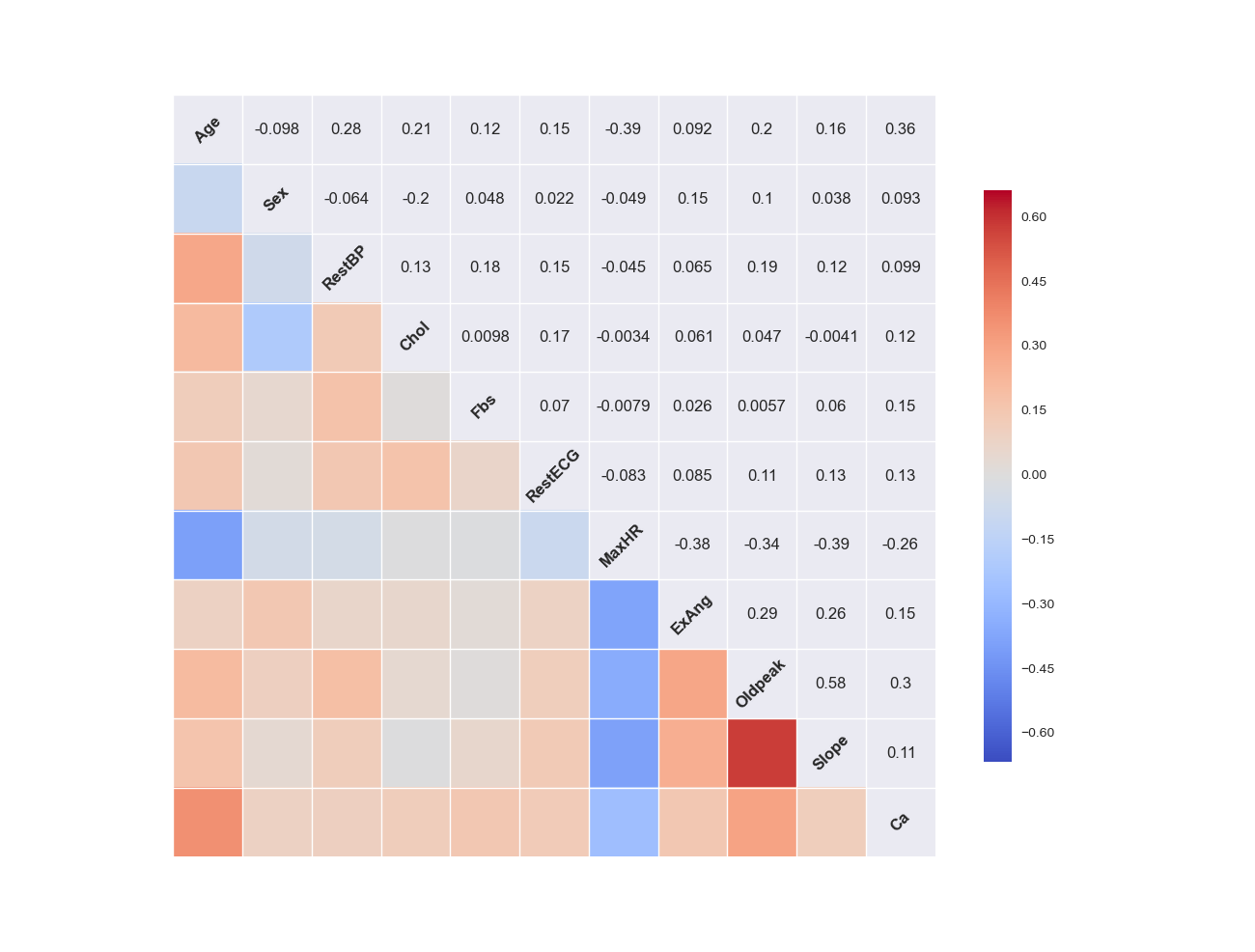
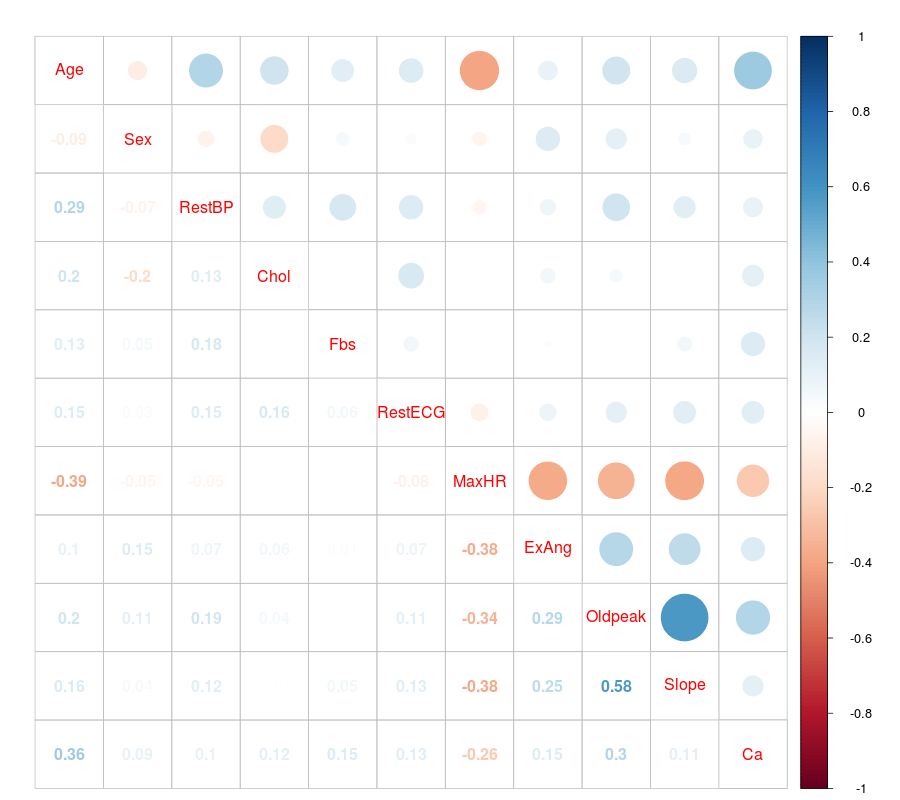
4.6.4. Correlation Matrix plot of the quantitative predictors¶
Python
- Correlation Matrix plot in Python
# cocorrelation Matrix plot
pd.DataFrame.corr(rawdata)
plt.show()
Then you will get get Figure Correlation Matrix plot in Python.
R
- Correlation Matrix plot in R
dev.off()
# laod cocorrelation Matrix plot lib
library(corrplot)
M <- cor(numdata)
#par(mfrow =c (1,2))
#corrplot(M, method = "square")
corrplot.mixed(M)
Then you will get Figure Correlation Matrix plot in R.
4.7. Source Code for This Section¶
The code for this section is available for download for R for Python,
Python
- Python Source code
'''
Created on Apr 25, 2016
test code
@author: Wenqiang Feng
'''
import pandas as pd
#import numpy as np
import matplotlib.pyplot as plt
from pandas.tools.plotting import scatter_matrix
from docutils.parsers.rst.directives import path
if __name__ == '__main__':
path ='~/Dropbox/MachineLearningAlgorithms/python_code/data/Heart.csv'
rawdata = pd.read_csv(path)
print "data summary"
print rawdata.describe()
# summary plot of the data
scatter_matrix(rawdata,figsize=[15,15])
plt.show()
# Histogram
rawdata.hist()
plt.show()
# boxplot
pd.DataFrame.boxplot(rawdata)
plt.show()
print "Raw data size"
nrow, ncol = rawdata.shape
print nrow, ncol
path = ('/home/feng/Dropbox/MachineLearningAlgorithms/python_code/data/'
'energy_efficiency.xlsx')
path
rawdataEnergy= pd.read_excel(path,sheetname=0)
nrow=rawdata.shape[0] #gives number of row count
ncol=rawdata.shape[1] #gives number of col count
print nrow, ncol
col_names = rawdata.columns.tolist()
print "Column names:"
print col_names
print "Data Format:"
print rawdata.dtypes
print "\nSample data:"
print(rawdata.head(6))
print "\n correlation Matrix"
print rawdata.corr()
# cocorrelation Matrix plot
pd.DataFrame.corr(rawdata)
plt.show()
print "\n covariance Matrix"
print rawdata.cov()
print rawdata[['Age','Ca']].corr()
pd.DataFrame.corr(rawdata)
plt.show()
# define colors list, to be used to plot survived either red (=0) or green (=1)
colors=['red','green']
# make a scatter plot
# rawdata.info()
from scipy import stats
import seaborn as sns # just a conventional alias, don't know why
sns.corrplot(rawdata) # compute and plot the pair-wise correlations
# save to file, remove the big white borders
#plt.savefig('attribute_correlations.png', tight_layout=True)
plt.show()
attr = rawdata['Age']
sns.distplot(attr)
plt.show()
sns.distplot(attr, kde=False, fit=stats.gamma);
plt.show()
# Two subplots, the axes array is 1-d
plt.figure(1)
plt.title('Histogram of Age')
plt.subplot(211) # 21,1 means first one of 2 rows, 1 col
sns.distplot(attr)
plt.subplot(212) # 21,2 means second one of 2 rows, 1 col
sns.distplot(attr, kde=False, fit=stats.gamma);
plt.show()
R
- R Source code
rm(list = ls())
# set the enverionment
path ='~/Dropbox/MachineLearningAlgorithms/python_code/data/Heart.csv'
rawdata = read.csv(path)
# summary of the data
summary(rawdata)
# plot of the summary
plot(rawdata)
dim(rawdata)
head(rawdata)
tail(rawdata)
colnames(rawdata)
attach(rawdata)
# get numerical data and remove NAN
numdata=na.omit(rawdata[,c(1:2,4:12)])
cor(numdata)
cov(numdata)
dev.off()
# laod cocorrelation Matrix plot lib
library(corrplot)
M <- cor(numdata)
#par(mfrow =c (1,2))
#corrplot(M, method = "square")
corrplot.mixed(M)
nrow=nrow(rawdata)
ncol=ncol(rawdata)
c(nrow, ncol)
Nvars=ncol(numdata)
# checking data format
typeof(rawdata)
install.packages("mlbench")
library(mlbench)
sapply(rawdata, class)
dev.off()
name=colnames(numdata)
Nvars=ncol(numdata)
# boxplot
par(mfrow =c (4,3))
for (i in 1:Nvars)
{
#boxplot(numdata[,i]~numdata[,Nvars],data=data,main=name[i])
boxplot(numdata[,i],data=numdata,main=name[i])
}
# Histogram with normal curve plot
dev.off()
Nvars=ncol(numdata)
name=colnames(numdata)
par(mfrow =c (3,5))
for (i in 1:Nvars)
{
x<- numdata[,i]
h<-hist(x, breaks=10, freq=TRUE, col="blue", xlab=name[i],main=" ",
font.lab=1)
axis(1, tck=1, col.ticks="light gray")
axis(1, tck=-0.015, col.ticks="black")
axis(2, tck=1, col.ticks="light gray", lwd.ticks="1")
axis(2, tck=-0.015)
xfit<-seq(min(x),max(x),length=40)
yfit<-dnorm(xfit,mean=mean(x),sd=sd(x))
yfit <- yfit*diff(h$mids[1:2])*length(x)
lines(xfit, yfit, col="blue", lwd=2)
}
library(reshape2)
library(ggplot2)
d <- melt(diamonds[,-c(2:4)])
ggplot(d,aes(x = value)) +
facet_wrap(~variable,scales = "free_x") +
geom_histogram()