3. PySpark Data Audit Functions¶
3.1. Basic Functions¶
3.1.1. mkdir¶
-
PySparkAudit.PySparkAudit.mkdir(path)[source] Make a new directory. if it’s exist, keep the old files.
Parameters: path – the directory path
3.1.2. mkdir_clean¶
-
PySparkAudit.PySparkAudit.mkdir_clean(path)[source] Make a new directory. if it’s exist, remove the old files.
Parameters: path – the directory path
3.1.3. df_merge¶
-
PySparkAudit.PySparkAudit.df_merge(dfs, key, how='left')[source] Merge multiple pandas data frames with same key.
Parameters: - dfs – name list of the data frames
- key – key for join
- how – method for join, the default value is left
Returns: merged data frame
3.1.4. data_types¶
-
PySparkAudit.PySparkAudit.data_types(df_in, tracking=False)[source] Generate the data types of the rdd data frame.
Parameters: - df_in – the input rdd data frame
- tracking – the flag for displaying CPU time, the default value is False
Returns: data types pandas data frame
>>> test = spark.createDataFrame([ ('Joe', 67, 'F', 7000, 'asymptomatic', 286.1, '2019-6-28'), ('Henry', 67, 'M', 8000, 'asymptomatic', 229.2, '2019-6-29'), ('Sam', 37, 'F', 6000, 'nonanginal', 250.3, '2019-6-30'), ('Max', 56, 'M', 9000, 'nontypical', 236.4, '2019-5-28'), ('Mat', 56, 'F', 9000, 'asymptomatic', 254.5, '2019-4-28')], ['Name', 'Age', 'Sex', 'Salary', 'ChestPain', 'Chol', 'CreatDate'] ) >>> test = test.withColumn('CreatDate', F.col('CreatDate').cast('timestamp')) >>> from PySparkAudit import data_types >>> data_types(test) feature dtypes 0 Name string 1 Age bigint 2 Sex string 3 Salary bigint 4 ChestPain string 5 Chol double 6 CreatDate timestamp
3.1.5. dtypes_class¶
-
PySparkAudit.PySparkAudit.dtypes_class(df_in)[source] Generate the data type categories: numerical, categorical, date and unsupported category.
Parameters: df_in – the input rdd data frame Returns: data type categories >>> test = spark.createDataFrame([ ('Joe', 67, 'F', 7000, 'asymptomatic', 286.1, '2019-6-28'), ('Henry', 67, 'M', 8000, 'asymptomatic', 229.2, '2019-6-29'), ('Sam', 37, 'F', 6000, 'nonanginal', 250.3, '2019-6-30'), ('Max', 56, 'M', 9000, 'nontypical', 236.4, '2019-5-28'), ('Mat', 56, 'F', 9000, 'asymptomatic', 254.5, '2019-4-28')], ['Name', 'Age', 'Sex', 'Salary', 'ChestPain', 'Chol', 'CreatDate'] ) >>> test = test.withColumn('CreatDate', F.col('CreatDate').cast('timestamp')) >>> from PySparkAudit import dtypes_class >>> dtypes_class(test) ( feature DataType 0 Name StringType 1 Age LongType 2 Sex StringType 3 Salary LongType 4 ChestPain StringType 5 Chol DoubleType 6 CreatDate TimestampType, ['Age', 'Salary', 'Chol'], ['Name', 'Sex', 'ChestPain'], ['CreatDate'], [])
3.1.6. counts¶
-
PySparkAudit.PySparkAudit.counts(df_in, tracking=False)[source] Generate the row counts and not null rows and distinct counts for each feature.
Parameters: - df_in – the input rdd data frame
- tracking – the flag for displaying CPU time, the default value is False
Returns: the counts in pandas data frame
>>> test = spark.createDataFrame([ ('Joe', None, 'F', 70000, 'asymptomatic', 286.1, '2019-6-28'), ('Henry', 67, 'M', 80000, 'asymptomatic', 229.2, '2019-6-29'), ('Sam', 37, 'F', 60000, 'nonanginal', 250.3, '2019-6-30'), ('Max', 56, ' ', 90000, None, 236.4, '2019-5-28'), ('Mat', 56, 'F', None, 'asymptomatic', 254.5, '2019-4-28')], ['Name', 'Age', 'Sex', 'Salary', 'ChestPain', 'Chol', 'CreatDate'] ) >>> test = test.withColumn('CreatDate', F.col('CreatDate').cast('timestamp')) >>> from PySparkAudit import counts >>> counts(test) feature row_count notnull_count distinct_count 0 Name 5 5 5 1 Age 5 4 3 2 Sex 5 5 3 3 Salary 5 4 4 4 ChestPain 5 4 2 5 Chol 5 5 5 6 CreatDate 5 5 5
3.1.7. describe¶
-
PySparkAudit.PySparkAudit.describe(df_in, columns=None, tracking=False)[source] Generate the simple data frame description using
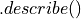 function in pyspark.
function in pyspark.Parameters: - df_in – the input rdd data frame
- columns – the specific feature columns, the default value is None
- tracking – the flag for displaying CPU time, the default value is False
Returns: the description in pandas data frame
>>> test = spark.createDataFrame([ ('Joe', 67, 'F', 7000, 'asymptomatic', 286.1, '2019-6-28'), ('Henry', 67, 'M', 8000, 'asymptomatic', 229.2, '2019-6-29'), ('Sam', 37, 'F', 6000, 'nonanginal', 250.3, '2019-6-30'), ('Max', 56, 'M', 9000, 'nontypical', 236.4, '2019-5-28'), ('Mat', 56, 'F', 9000, 'asymptomatic', 254.5, '2019-4-28')], ['Name', 'Age', 'Sex', 'Salary', 'ChestPain', 'Chol', 'CreatDate'] ) >>> test = test.withColumn('CreatDate', F.col('CreatDate').cast('timestamp')) >>> from PySparkAudit import describe >>> describe(test) summary count mean ... min max feature ... Name 5 None ... Henry Sam Age 5 56.6 ... 37 67 Sex 5 None ... F M Salary 5 78000.0 ... 60000 90000 ChestPain 5 None ... asymptomatic nontypical Chol 5 251.3 ... 229.2 286.1 CreatDate 5 None ... 2019-4-28 2019-6-30
[7 rows x 5 columns]
3.1.8. percentiles¶
-
PySparkAudit.PySparkAudit.percentiles(df_in, deciles=False, tracking=False)[source] Generate the percentiles for rdd data frame.
Parameters: - df_in – the input rdd data frame
- deciles – the flag for generate the deciles
- tracking – the flag for displaying CPU time, the default value is False
Returns: percentiles in pandas data frame
>>> test = spark.createDataFrame([ ('Joe', 67, 'F', 7000, 'asymptomatic', 286.1, '2019-6-28'), ('Henry', 67, 'M', 8000, 'asymptomatic', 229.2, '2019-6-29'), ('Sam', 37, 'F', 6000, 'nonanginal', 250.3, '2019-6-30'), ('Max', 56, 'M', 9000, 'nontypical', 236.4, '2019-5-28'), ('Mat', 56, 'F', 9000, 'asymptomatic', 254.5, '2019-4-28')], ['Name', 'Age', 'Sex', 'Salary', 'ChestPain', 'Chol', 'CreatDate'] ) >>> from PySparkAudit import percentiles >>> percentiles(test) feature Q1 Med Q3 0 Age 56.0 67.0 67.0 1 Salary 80000.0 90000.0 90000.0 2 Chol 250.3 254.5 286.1
3.1.9. feature_len¶
-
PySparkAudit.PySparkAudit.feature_len(df_in, tracking=False)[source] Generate feature length statistical results for each feature in the rdd data frame.
Parameters: - df_in – the input rdd data frame
- tracking – the flag for displaying CPU time, the default value is False
Returns: the feature length statistical results in pandas data frame
>>> test = spark.createDataFrame([ ('Joe', 67, 'F', 7000, 'asymptomatic', 286.1, '2019-6-28'), ('Henry', 67, 'M', 8000, 'asymptomatic', 229.2, '2019-6-29'), ('Sam', 37, 'F', 6000, 'nonanginal', 250.3, '2019-6-30'), ('Max', 56, 'M', 9000, 'nontypical', 236.4, '2019-5-28'), ('Mat', 56, 'F', 9000, 'asymptomatic', 254.5, '2019-4-28')], ['Name', 'Age', 'Sex', 'Salary', 'ChestPain', 'Chol', 'CreatDate'] ) >>> from PySparkAudit import feature_len >>> feature_len(test) feature min_length avg_length max_length 0 Name 3.0 3.4 5.0 1 Age 2.0 2.0 2.0 2 Sex 1.0 1.0 1.0 3 Salary 5.0 5.0 5.0 4 ChestPain 10.0 11.2 12.0 5 Chol 5.0 5.0 5.0 6 CreatDate 9.0 9.0 9.0
3.1.10. freq_items¶
-
PySparkAudit.PySparkAudit.freq_items(df_in, top_n=5, tracking=False)[source] Generate the top_n frequent items in for each feature in the rdd data frame.
Parameters: - df_in – the input rdd data frame
- top_n – the number of the most frequent item
- tracking – the flag for displaying CPU time, the default value is False
Returns: >>> test = spark.createDataFrame([ ('Joe', 67, 'F', 7000, 'asymptomatic', 286.1, '2019-6-28'), ('Henry', 67, 'M', 8000, 'asymptomatic', 229.2, '2019-6-29'), ('Sam', 37, 'F', 6000, 'nonanginal', 250.3, '2019-6-30'), ('Max', 56, 'M', 9000, 'nontypical', 236.4, '2019-5-28'), ('Mat', 56, 'F', 9000, 'asymptomatic', 254.5, '2019-4-28')], ['Name', 'Age', 'Sex', 'Salary', 'ChestPain', 'Chol', 'CreatDate'] ) >>> from PySparkAudit import freq_items >>> freq_items(test) feature freq_items[value, freq] 0 Name [[Joe, 1], [Mat, 1], [Henry, 1], [Sam, 1], [Ma... 1 Age [[67, 2], [56, 2], [37, 1]] 2 Sex [[F, 3], [M, 2]] 3 Salary [[90000, 2], [70000, 1], [80000, 1], [60000, 1]] 4 ChestPain [[asymptomatic, 3], [nontypical, 1], [nonangin... 5 Chol [[286.1, 1], [250.3, 1], [229.2, 1], [236.4, 1... 6 CreatDate [[2019-6-30, 1], [2019-5-28, 1], [2019-4-28, 1...
3.1.11. rates¶
-
PySparkAudit.PySparkAudit.rates(df_in, columns=None, numeric=True, tracking=False)[source] Generate the null, empty, negative, zero and positive value rates and feature variance for each feature in the rdd data frame.
Parameters: - df_in – the input rdd data frame
- columns – the specific feature columns, the default value is None
- numeric – the flag for numerical rdd data frame, the default value is True
- tracking – the flag for displaying CPU time, the default value is False
Returns: the null, empty, negative, zero and positive value rates and feature variance in pandas data frame
>>> test = spark.createDataFrame([ ('Joe', 67, 'F', 7000, 'asymptomatic', 286.1, '2019-6-28'), ('Henry', 67, 'M', 8000, 'asymptomatic', 229.2, '2019-6-29'), ('Sam', 37, 'F', 6000, 'nonanginal', 250.3, '2019-6-30'), ('Max', 56, 'M', 9000, 'nontypical', 236.4, '2019-5-28'), ('Mat', 56, 'F', 9000, 'asymptomatic', 254.5, '2019-4-28')], ['Name', 'Age', 'Sex', 'Salary', 'ChestPain', 'Chol', 'CreatDate'] ) >>> from PySparkAudit import rates >>> rates(test) feature feature_variance ... rate_zero rate_pos 0 Age 0.6 ... 0.0 1.0 1 Salary 0.8 ... 0.0 1.0 2 Chol 1.0 ... 0.0 1.0 3 Name 1.0 ... 0.0 0.0 4 Sex 0.4 ... 0.0 0.0 5 ChestPain 0.6 ... 0.0 0.0 6 CreatDate 1.0 ... 0.0 0.0
[7 rows x 7 columns]
3.1.12. corr_matrix¶
-
PySparkAudit.PySparkAudit.corr_matrix(df_in, method='pearson', output_dir=None, rotation=True, display=False, tracking=False)[source] Generate the correlation matrix and heat map plot for rdd data frame.
Parameters: - df_in – the input rdd data frame
- method – the method which applied to calculate the correlation matrix: pearson or spearman. the default value is pearson
- output_dir – the out put directory, the default value is the current working directory
- rotation – the flag for rotating the xticks in the plot, the default value is True
- display – the flag for displaying the figures, the default value is False
- tracking – the flag for displaying CPU time, the default value is False
Returns: the correlation matrix in pandas data frame
>>> test = spark.createDataFrame([ ('Joe', 67, 'F', 7000, 'asymptomatic', 286.1, '2019-6-28'), ('Henry', 67, 'M', 8000, 'asymptomatic', 229.2, '2019-6-29'), ('Sam', 37, 'F', 6000, 'nonanginal', 250.3, '2019-6-30'), ('Max', 56, 'M', 9000, 'nontypical', 236.4, '2019-5-28'), ('Mat', 56, 'F', 9000, 'asymptomatic', 254.5, '2019-4-28')], ['Name', 'Age', 'Sex', 'Salary', 'ChestPain', 'Chol', 'CreatDate'] ) >>> from PySparkAudit import corr_matrix >>> corr_matrix(test) ================================================================ The correlation matrix plot Corr.png was located at: /home/feng/Audited Age Salary Chol Age 1.000000 0.431663 0.147226 Salary 0.431663 1.000000 -0.388171 Chol 0.147226 -0.388171 1.000000
3.2. Plot Functions¶
3.2.1. hist_plot¶
-
PySparkAudit.PySparkAudit.hist_plot(df_in, bins=50, output_dir=None, sample_size=None, display=False, tracking=False)[source] Histogram plot for the numerical features in the rdd data frame. This part is super time and memory consuming. If the data size is larger than 10,000, the histograms will be saved in .pdf format. Otherwise, the histograms will be saved in .png format in hist folder.
If your time and memory are limited, you can use sample_size to generate the subset of the data frame to generate the histograms.
Parameters: - df_in – the input rdd data frame
- bins – the number of bins for generate the bar plots
- output_dir – the out put directory, the default value is the current working directory
- sample_size – the size for generate the subset from the rdd data frame, the default value none
- display – the flag for displaying the figures, the default value is False
- tracking – the flag for displaying CPU time, the default value is False
3.2.2. bar_plot¶
-
PySparkAudit.PySparkAudit.bar_plot(df_in, top_n=20, rotation=True, output_dir=None, display=False, tracking=False)[source] Bar plot for the categorical features in the rdd data frame.
Parameters: - df_in – the input rdd data frame
- top_n – the number of the most frequent feature to show in the bar plot
- rotation – the flag for rotating the xticks in the plot, the default value is True
- output_dir – the out put directory, the default value is the current working directory
- display – the flag for displaying the figures, the default value is False
- tracking – the flag for displaying CPU time, the default value is False
3.2.3. trend_plot¶
-
PySparkAudit.PySparkAudit.trend_plot(df_in, types='day', d_time=None, rotation=True, output_dir=None, display=False, tracking=False)[source] Trend plot for the aggregated time series data if the rdd data frame has date features and numerical features.
Parameters: - df_in – the input rdd data frame
- types – the types for time feature aggregation: day, month, year, the default value is day
- d_time – the specific feature name of the date feature, the default value is the first date feature in the rdd data frame
- rotation – the flag for rotating the xticks in the plot, the default value is True
- output_dir – the out put directory, the default value is the current working directory
- display – the flag for displaying the figures, the default value is False
- tracking – the flag for displaying CPU time, the default value is False
3.3. Summary Functions¶
3.3.1. dataset_summary¶
-
PySparkAudit.PySparkAudit.dataset_summary(df_in, tracking=False)[source] The data set basics summary.
Parameters: - df_in – the input rdd data frame
- tracking – the flag for displaying CPU time, the default value is False
Returns: data set summary in pandas data frame
3.3.2. numeric_summary¶
-
PySparkAudit.PySparkAudit.numeric_summary(df_in, columns=None, deciles=False, top_n=5, tracking=False)[source] The auditing function for numerical rdd data frame.
Parameters: - df_in – the input rdd data frame
- columns – the specific feature columns, the default value is None
- deciles – the flag for generate the deciles
- top_n – the number of the most frequent item
- tracking – the flag for displaying CPU time, the default value is False
Returns: the audited results for the numerical features in pandas data frame
3.3.3. category_summary¶
-
PySparkAudit.PySparkAudit.category_summary(df_in, columns=None, top_n=5, tracking=False)[source] The auditing function for categorical rdd data frame.
Parameters: - df_in – the input rdd data frame
- columns – the specific feature columns, the default value is None
- top_n – the number of the most frequent item
- tracking – the flag for displaying CPU time, the default value is False
Returns: the audited results for the categorical features in pandas data frame
3.4. Auditing Function¶
3.4.1. auditing¶
-
PySparkAudit.PySparkAudit.auditing(df_in, writer=None, columns=None, deciles=False, top_freq_item=5, bins=50, top_cat_item=20, method='pearson', output_dir=None, types='day', d_time=None, rotation=True, sample_size=None, display=False, tracking=False)[source] The wrapper of auditing functions.
Parameters: - df_in – the input rdd data frame
- writer – the writer for excel output
- columns – the specific feature columns, the default value is None
- deciles – the flag for generate the deciles
- top_freq_item – the number of the most frequent item
- bins – the number of bins for generate the bar plots
- top_cat_item – the number of the most frequent feature to show in the bar plot
- method – the method which applied to calculate the correlation matrix: pearson or spearman. the default value is pearson
- output_dir – the out put directory, the default value is the current working directory
- types – the types for time feature aggregation: day, month, year, the default value is day
- d_time – the specific feature name of the date feature, the default value is the first date feature in the rdd data frame
- rotation – the flag for rotating the xticks in the plot, the default value is True
- sample_size – the size for generate the subset from the rdd data frame, the default value none
- display – the flag for displaying the figures, the default value is False
- tracking – the flag for displaying CPU time, the default value is False
Returns: the all audited results in pandas data frame
3.5. Plotting Function¶
3.5.1. fig_plots¶
-
PySparkAudit.PySparkAudit.fig_plots(df_in, output_dir=None, bins=50, top_n=20, types='day', d_time=None, rotation=True, sample_size=None, display=False, tracking=False)[source] The wrapper for the plot functions.
Parameters: - df_in – the input rdd data frame
- output_dir – the out put directory, the default value is the current working directory
- bins – the number of bins for generate the bar plots
- top_n – the number of the most frequent feature to show in the bar plot
- types – the types for time feature aggregation: day, month, year, the default value is day
- d_time – the specific feature name of the date feature, the default value is the first date feature in the rdd data frame
- rotation – the flag for rotating the xticks in the plot, the default value is True
- sample_size – the size for generate the subset from the rdd data frame, the default value none
- display – the flag for displaying the figures, the default value is False
- tracking – the flag for displaying CPU time, the default value is False