27. PySpark API¶
Those APIs are automatically generated from PySpark package, so all the CopyRights belong to Spark.
27.1. Stat API¶
-
class
pyspark.ml.stat.ChiSquareTest[source]¶ Note
Experimental
Conduct Pearson’s independence test for every feature against the label. For each feature, the (feature, label) pairs are converted into a contingency matrix for which the Chi-squared statistic is computed. All label and feature values must be categorical.
The null hypothesis is that the occurrence of the outcomes is statistically independent.
New in version 2.2.0.
-
static
test(dataset, featuresCol, labelCol)[source]¶ Perform a Pearson’s independence test using dataset.
- Parameters
dataset – DataFrame of categorical labels and categorical features. Real-valued features will be treated as categorical for each distinct value.
featuresCol – Name of features column in dataset, of type
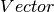 (
(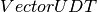 ).
).labelCol – Name of label column in dataset, of any numerical type.
- Returns
DataFrame containing the test result for every feature against the label. This DataFrame will contain a single Row with the following fields: -
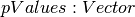 -
- ![degreesOfFreedom: Array[Int]](_images/math/b57379873ad0a331db8ba5752cafbe2f1ba8a6b5.png) -
- 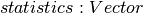 Each of these fields has one value per feature.
Each of these fields has one value per feature.
>>> from pyspark.ml.linalg import Vectors >>> from pyspark.ml.stat import ChiSquareTest >>> dataset = [[0, Vectors.dense([0, 0, 1])], ... [0, Vectors.dense([1, 0, 1])], ... [1, Vectors.dense([2, 1, 1])], ... [1, Vectors.dense([3, 1, 1])]] >>> dataset = spark.createDataFrame(dataset, ["label", "features"]) >>> chiSqResult = ChiSquareTest.test(dataset, 'features', 'label') >>> chiSqResult.select("degreesOfFreedom").collect()[0] Row(degreesOfFreedom=[3, 1, 0])
New in version 2.2.0.
-
static
-
class
pyspark.ml.stat.Correlation[source]¶ Note
Experimental
Compute the correlation matrix for the input dataset of Vectors using the specified method. Methods currently supported:
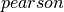 (default),
(default), 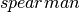 .
.Note
For Spearman, a rank correlation, we need to create an RDD[Double] for each column and sort it in order to retrieve the ranks and then join the columns back into an RDD[Vector], which is fairly costly. Cache the input Dataset before calling corr with
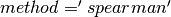 to avoid recomputing the common lineage.
to avoid recomputing the common lineage.New in version 2.2.0.
-
static
corr(dataset, column, method='pearson')[source]¶ Compute the correlation matrix with specified method using dataset.
- Parameters
dataset – A Dataset or a DataFrame.
column – The name of the column of vectors for which the correlation coefficient needs to be computed. This must be a column of the dataset, and it must contain Vector objects.
method – String specifying the method to use for computing correlation. Supported:
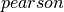 (default),
(default), 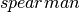 .
.
- Returns
A DataFrame that contains the correlation matrix of the column of vectors. This DataFrame contains a single row and a single column of name ‘$METHODNAME($COLUMN)’.
>>> from pyspark.ml.linalg import Vectors >>> from pyspark.ml.stat import Correlation >>> dataset = [[Vectors.dense([1, 0, 0, -2])], ... [Vectors.dense([4, 5, 0, 3])], ... [Vectors.dense([6, 7, 0, 8])], ... [Vectors.dense([9, 0, 0, 1])]] >>> dataset = spark.createDataFrame(dataset, ['features']) >>> pearsonCorr = Correlation.corr(dataset, 'features', 'pearson').collect()[0][0] >>> print(str(pearsonCorr).replace('nan', 'NaN')) DenseMatrix([[ 1. , 0.0556..., NaN, 0.4004...], [ 0.0556..., 1. , NaN, 0.9135...], [ NaN, NaN, 1. , NaN], [ 0.4004..., 0.9135..., NaN, 1. ]]) >>> spearmanCorr = Correlation.corr(dataset, 'features', method='spearman').collect()[0][0] >>> print(str(spearmanCorr).replace('nan', 'NaN')) DenseMatrix([[ 1. , 0.1054..., NaN, 0.4 ], [ 0.1054..., 1. , NaN, 0.9486... ], [ NaN, NaN, 1. , NaN], [ 0.4 , 0.9486... , NaN, 1. ]])
New in version 2.2.0.
-
static
-
class
pyspark.ml.stat.KolmogorovSmirnovTest[source]¶ Note
Experimental
Conduct the two-sided Kolmogorov Smirnov (KS) test for data sampled from a continuous distribution.
By comparing the largest difference between the empirical cumulative distribution of the sample data and the theoretical distribution we can provide a test for the the null hypothesis that the sample data comes from that theoretical distribution.
New in version 2.4.0.
-
static
test(dataset, sampleCol, distName, *params)[source]¶ Conduct a one-sample, two-sided Kolmogorov-Smirnov test for probability distribution equality. Currently supports the normal distribution, taking as parameters the mean and standard deviation.
- Parameters
dataset – a Dataset or a DataFrame containing the sample of data to test.
sampleCol – Name of sample column in dataset, of any numerical type.
distName – a
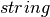 name for a theoretical distribution, currently only support “norm”.
name for a theoretical distribution, currently only support “norm”.params – a list of
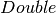 values specifying the parameters to be used for the theoretical
distribution. For “norm” distribution, the parameters includes mean and variance.
values specifying the parameters to be used for the theoretical
distribution. For “norm” distribution, the parameters includes mean and variance.
- Returns
A DataFrame that contains the Kolmogorov-Smirnov test result for the input sampled data. This DataFrame will contain a single Row with the following fields: -
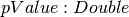 -
- 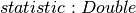
>>> from pyspark.ml.stat import KolmogorovSmirnovTest >>> dataset = [[-1.0], [0.0], [1.0]] >>> dataset = spark.createDataFrame(dataset, ['sample']) >>> ksResult = KolmogorovSmirnovTest.test(dataset, 'sample', 'norm', 0.0, 1.0).first() >>> round(ksResult.pValue, 3) 1.0 >>> round(ksResult.statistic, 3) 0.175 >>> dataset = [[2.0], [3.0], [4.0]] >>> dataset = spark.createDataFrame(dataset, ['sample']) >>> ksResult = KolmogorovSmirnovTest.test(dataset, 'sample', 'norm', 3.0, 1.0).first() >>> round(ksResult.pValue, 3) 1.0 >>> round(ksResult.statistic, 3) 0.175
New in version 2.4.0.
-
static
-
class
pyspark.ml.stat.Summarizer[source]¶ Note
Experimental
Tools for vectorized statistics on MLlib Vectors. The methods in this package provide various statistics for Vectors contained inside DataFrames. This class lets users pick the statistics they would like to extract for a given column.
>>> from pyspark.ml.stat import Summarizer >>> from pyspark.sql import Row >>> from pyspark.ml.linalg import Vectors >>> summarizer = Summarizer.metrics("mean", "count") >>> df = sc.parallelize([Row(weight=1.0, features=Vectors.dense(1.0, 1.0, 1.0)), ... Row(weight=0.0, features=Vectors.dense(1.0, 2.0, 3.0))]).toDF() >>> df.select(summarizer.summary(df.features, df.weight)).show(truncate=False) +-----------------------------------+ |aggregate_metrics(features, weight)| +-----------------------------------+ |[[1.0,1.0,1.0], 1] | +-----------------------------------+ >>> df.select(summarizer.summary(df.features)).show(truncate=False) +--------------------------------+ |aggregate_metrics(features, 1.0)| +--------------------------------+ |[[1.0,1.5,2.0], 2] | +--------------------------------+ >>> df.select(Summarizer.mean(df.features, df.weight)).show(truncate=False) +--------------+ |mean(features)| +--------------+ |[1.0,1.0,1.0] | +--------------+ >>> df.select(Summarizer.mean(df.features)).show(truncate=False) +--------------+ |mean(features)| +--------------+ |[1.0,1.5,2.0] | +--------------+
New in version 2.4.0.
-
static
metrics(*metrics)[source]¶ Given a list of metrics, provides a builder that it turns computes metrics from a column.
See the documentation of [[Summarizer]] for an example.
- The following metrics are accepted (case sensitive):
mean: a vector that contains the coefficient-wise mean.
variance: a vector tha contains the coefficient-wise variance.
count: the count of all vectors seen.
numNonzeros: a vector with the number of non-zeros for each coefficients
max: the maximum for each coefficient.
min: the minimum for each coefficient.
normL2: the Euclidian norm for each coefficient.
normL1: the L1 norm of each coefficient (sum of the absolute values).
- Parameters
metrics – metrics that can be provided.
- Returns
an object of
pyspark.ml.stat.SummaryBuilder
Note: Currently, the performance of this interface is about 2x~3x slower then using the RDD interface.
New in version 2.4.0.
-
static
-
class
pyspark.ml.stat.SummaryBuilder(jSummaryBuilder)[source]¶ Note
Experimental
A builder object that provides summary statistics about a given column.
Users should not directly create such builders, but instead use one of the methods in
pyspark.ml.stat.SummarizerNew in version 2.4.0.
-
summary(featuresCol, weightCol=None)[source]¶ Returns an aggregate object that contains the summary of the column with the requested metrics.
- Parameters
featuresCol – a column that contains features Vector object.
weightCol – a column that contains weight value. Default weight is 1.0.
- Returns
an aggregate column that contains the statistics. The exact content of this structure is determined during the creation of the builder.
New in version 2.4.0.
-
27.2. Regression API¶
-
class
pyspark.ml.regression.AFTSurvivalRegression(*args, **kwargs)[source]¶ Note
Experimental
Accelerated Failure Time (AFT) Model Survival Regression
Fit a parametric AFT survival regression model based on the Weibull distribution of the survival time.
See also
>>> from pyspark.ml.linalg import Vectors >>> df = spark.createDataFrame([ ... (1.0, Vectors.dense(1.0), 1.0), ... (1e-40, Vectors.sparse(1, [], []), 0.0)], ["label", "features", "censor"]) >>> aftsr = AFTSurvivalRegression() >>> model = aftsr.fit(df) >>> model.predict(Vectors.dense(6.3)) 1.0 >>> model.predictQuantiles(Vectors.dense(6.3)) DenseVector([0.0101, 0.0513, 0.1054, 0.2877, 0.6931, 1.3863, 2.3026, 2.9957, 4.6052]) >>> model.transform(df).show() +-------+---------+------+----------+ | label| features|censor|prediction| +-------+---------+------+----------+ | 1.0| [1.0]| 1.0| 1.0| |1.0E-40|(1,[],[])| 0.0| 1.0| +-------+---------+------+----------+ ... >>> aftsr_path = temp_path + "/aftsr" >>> aftsr.save(aftsr_path) >>> aftsr2 = AFTSurvivalRegression.load(aftsr_path) >>> aftsr2.getMaxIter() 100 >>> model_path = temp_path + "/aftsr_model" >>> model.save(model_path) >>> model2 = AFTSurvivalRegressionModel.load(model_path) >>> model.coefficients == model2.coefficients True >>> model.intercept == model2.intercept True >>> model.scale == model2.scale True
New in version 1.6.0.
-
getQuantileProbabilities()[source]¶ Gets the value of quantileProbabilities or its default value.
New in version 1.6.0.
-
getQuantilesCol()[source]¶ Gets the value of quantilesCol or its default value.
New in version 1.6.0.
-
setParams(featuresCol='features', labelCol='label', predictionCol='prediction', fitIntercept=True, maxIter=100, tol=1e-06, censorCol='censor', quantileProbabilities=[0.01, 0.05, 0.1, 0.25, 0.5, 0.75, 0.9, 0.95, 0.99], quantilesCol=None, aggregationDepth=2)[source]¶ setParams(self, featuresCol=”features”, labelCol=”label”, predictionCol=”prediction”, fitIntercept=True, maxIter=100, tol=1E-6, censorCol=”censor”, quantileProbabilities=[0.01, 0.05, 0.1, 0.25, 0.5, 0.75, 0.9, 0.95, 0.99], quantilesCol=None, aggregationDepth=2):
New in version 1.6.0.
-
-
class
pyspark.ml.regression.AFTSurvivalRegressionModel(java_model=None)[source]¶ Note
Experimental
Model fitted by
AFTSurvivalRegression.New in version 1.6.0.
-
class
pyspark.ml.regression.DecisionTreeRegressionModel(java_model=None)[source]¶ Model fitted by
DecisionTreeRegressor.New in version 1.4.0.
-
property
featureImportances[source]¶ Estimate of the importance of each feature.
This generalizes the idea of “Gini” importance to other losses, following the explanation of Gini importance from “Random Forests” documentation by Leo Breiman and Adele Cutler, and following the implementation from scikit-learn.
- This feature importance is calculated as follows:
importance(feature j) = sum (over nodes which split on feature j) of the gain, where gain is scaled by the number of instances passing through node
Normalize importances for tree to sum to 1.
Note
Feature importance for single decision trees can have high variance due to correlated predictor variables. Consider using a
RandomForestRegressorto determine feature importance instead.New in version 2.0.0.
-
property
-
class
pyspark.ml.regression.DecisionTreeRegressor(*args, **kwargs)[source]¶ Decision tree learning algorithm for regression. It supports both continuous and categorical features.
>>> from pyspark.ml.linalg import Vectors >>> df = spark.createDataFrame([ ... (1.0, Vectors.dense(1.0)), ... (0.0, Vectors.sparse(1, [], []))], ["label", "features"]) >>> dt = DecisionTreeRegressor(maxDepth=2, varianceCol="variance") >>> model = dt.fit(df) >>> model.depth 1 >>> model.numNodes 3 >>> model.featureImportances SparseVector(1, {0: 1.0}) >>> model.numFeatures 1 >>> test0 = spark.createDataFrame([(Vectors.dense(-1.0),)], ["features"]) >>> model.transform(test0).head().prediction 0.0 >>> test1 = spark.createDataFrame([(Vectors.sparse(1, [0], [1.0]),)], ["features"]) >>> model.transform(test1).head().prediction 1.0 >>> dtr_path = temp_path + "/dtr" >>> dt.save(dtr_path) >>> dt2 = DecisionTreeRegressor.load(dtr_path) >>> dt2.getMaxDepth() 2 >>> model_path = temp_path + "/dtr_model" >>> model.save(model_path) >>> model2 = DecisionTreeRegressionModel.load(model_path) >>> model.numNodes == model2.numNodes True >>> model.depth == model2.depth True >>> model.transform(test1).head().variance 0.0
New in version 1.4.0.
-
setParams(self, featuresCol='features', labelCol='label', predictionCol='prediction', maxDepth=5, maxBins=32, minInstancesPerNode=1, minInfoGain=0.0, maxMemoryInMB=256, cacheNodeIds=False, checkpointInterval=10, impurity='variance', seed=None, varianceCol=None)[source]¶ Sets params for the DecisionTreeRegressor.
New in version 1.4.0.
-
-
class
pyspark.ml.regression.GBTRegressionModel(java_model=None)[source]¶ Model fitted by
GBTRegressor.New in version 1.4.0.
-
evaluateEachIteration(dataset, loss)[source]¶ Method to compute error or loss for every iteration of gradient boosting.
- Parameters
dataset – Test dataset to evaluate model on, where dataset is an instance of
pyspark.sql.DataFrameloss – The loss function used to compute error. Supported options: squared, absolute
New in version 2.4.0.
-
property
featureImportances[source]¶ Estimate of the importance of each feature.
Each feature’s importance is the average of its importance across all trees in the ensemble The importance vector is normalized to sum to 1. This method is suggested by Hastie et al. (Hastie, Tibshirani, Friedman. “The Elements of Statistical Learning, 2nd Edition.” 2001.) and follows the implementation from scikit-learn.
New in version 2.0.0.
-
-
class
pyspark.ml.regression.GBTRegressor(*args, **kwargs)[source]¶ Gradient-Boosted Trees (GBTs) learning algorithm for regression. It supports both continuous and categorical features.
>>> from numpy import allclose >>> from pyspark.ml.linalg import Vectors >>> df = spark.createDataFrame([ ... (1.0, Vectors.dense(1.0)), ... (0.0, Vectors.sparse(1, [], []))], ["label", "features"]) >>> gbt = GBTRegressor(maxIter=5, maxDepth=2, seed=42) >>> print(gbt.getImpurity()) variance >>> print(gbt.getFeatureSubsetStrategy()) all >>> model = gbt.fit(df) >>> model.featureImportances SparseVector(1, {0: 1.0}) >>> model.numFeatures 1 >>> allclose(model.treeWeights, [1.0, 0.1, 0.1, 0.1, 0.1]) True >>> test0 = spark.createDataFrame([(Vectors.dense(-1.0),)], ["features"]) >>> model.transform(test0).head().prediction 0.0 >>> test1 = spark.createDataFrame([(Vectors.sparse(1, [0], [1.0]),)], ["features"]) >>> model.transform(test1).head().prediction 1.0 >>> gbtr_path = temp_path + "gbtr" >>> gbt.save(gbtr_path) >>> gbt2 = GBTRegressor.load(gbtr_path) >>> gbt2.getMaxDepth() 2 >>> model_path = temp_path + "gbtr_model" >>> model.save(model_path) >>> model2 = GBTRegressionModel.load(model_path) >>> model.featureImportances == model2.featureImportances True >>> model.treeWeights == model2.treeWeights True >>> model.trees [DecisionTreeRegressionModel (uid=...) of depth..., DecisionTreeRegressionModel...] >>> validation = spark.createDataFrame([(0.0, Vectors.dense(-1.0))], ... ["label", "features"]) >>> model.evaluateEachIteration(validation, "squared") [0.0, 0.0, 0.0, 0.0, 0.0]
New in version 1.4.0.
-
setFeatureSubsetStrategy(value)[source]¶ Sets the value of
featureSubsetStrategy.New in version 2.4.0.
-
setParams(self, featuresCol='features', labelCol='label', predictionCol='prediction', maxDepth=5, maxBins=32, minInstancesPerNode=1, minInfoGain=0.0, maxMemoryInMB=256, cacheNodeIds=False, subsamplingRate=1.0, checkpointInterval=10, lossType='squared', maxIter=20, stepSize=0.1, seed=None, impurity='variance', featureSubsetStrategy='all')[source]¶ Sets params for Gradient Boosted Tree Regression.
New in version 1.4.0.
-
-
class
pyspark.ml.regression.GeneralizedLinearRegression(*args, **kwargs)[source]¶ Note
Experimental
Generalized Linear Regression.
Fit a Generalized Linear Model specified by giving a symbolic description of the linear predictor (link function) and a description of the error distribution (family). It supports “gaussian”, “binomial”, “poisson”, “gamma” and “tweedie” as family. Valid link functions for each family is listed below. The first link function of each family is the default one.
“gaussian” -> “identity”, “log”, “inverse”
“binomial” -> “logit”, “probit”, “cloglog”
“poisson” -> “log”, “identity”, “sqrt”
“gamma” -> “inverse”, “identity”, “log”
“tweedie” -> power link function specified through “linkPower”. The default link power in the tweedie family is 1 - variancePower.
See also
>>> from pyspark.ml.linalg import Vectors >>> df = spark.createDataFrame([ ... (1.0, Vectors.dense(0.0, 0.0)), ... (1.0, Vectors.dense(1.0, 2.0)), ... (2.0, Vectors.dense(0.0, 0.0)), ... (2.0, Vectors.dense(1.0, 1.0)),], ["label", "features"]) >>> glr = GeneralizedLinearRegression(family="gaussian", link="identity", linkPredictionCol="p") >>> model = glr.fit(df) >>> transformed = model.transform(df) >>> abs(transformed.head().prediction - 1.5) < 0.001 True >>> abs(transformed.head().p - 1.5) < 0.001 True >>> model.coefficients DenseVector([1.5..., -1.0...]) >>> model.numFeatures 2 >>> abs(model.intercept - 1.5) < 0.001 True >>> glr_path = temp_path + "/glr" >>> glr.save(glr_path) >>> glr2 = GeneralizedLinearRegression.load(glr_path) >>> glr.getFamily() == glr2.getFamily() True >>> model_path = temp_path + "/glr_model" >>> model.save(model_path) >>> model2 = GeneralizedLinearRegressionModel.load(model_path) >>> model.intercept == model2.intercept True >>> model.coefficients[0] == model2.coefficients[0] True
New in version 2.0.0.
-
getLinkPredictionCol()[source]¶ Gets the value of linkPredictionCol or its default value.
New in version 2.0.0.
-
getVariancePower()[source]¶ Gets the value of variancePower or its default value.
New in version 2.2.0.
-
setParams(self, labelCol='label', featuresCol='features', predictionCol='prediction', family='gaussian', link=None, fitIntercept=True, maxIter=25, tol=1e-06, regParam=0.0, weightCol=None, solver='irls', linkPredictionCol=None, variancePower=0.0, linkPower=None, offsetCol=None)[source]¶ Sets params for generalized linear regression.
New in version 2.0.0.
-
class
pyspark.ml.regression.GeneralizedLinearRegressionModel(java_model=None)[source]¶ Note
Experimental
Model fitted by
GeneralizedLinearRegression.New in version 2.0.0.
-
evaluate(dataset)[source]¶ Evaluates the model on a test dataset.
- Parameters
dataset – Test dataset to evaluate model on, where dataset is an instance of
pyspark.sql.DataFrame
New in version 2.0.0.
-
-
class
pyspark.ml.regression.GeneralizedLinearRegressionSummary(java_obj=None)[source]¶ Note
Experimental
Generalized linear regression results evaluated on a dataset.
New in version 2.0.0.
-
property
aic[source]¶ Akaike’s “An Information Criterion”(AIC) for the fitted model.
New in version 2.0.0.
-
property
dispersion[source]¶ The dispersion of the fitted model. It is taken as 1.0 for the “binomial” and “poisson” families, and otherwise estimated by the residual Pearson’s Chi-Squared statistic (which is defined as sum of the squares of the Pearson residuals) divided by the residual degrees of freedom.
New in version 2.0.0.
-
property
predictionCol[source]¶ Field in
predictionswhich gives the predicted value of each instance. This is set to a new column name if the original model’s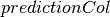 is not set.
is not set.New in version 2.0.0.
-
property
-
class
pyspark.ml.regression.GeneralizedLinearRegressionTrainingSummary(java_obj=None)[source]¶ Note
Experimental
Generalized linear regression training results.
New in version 2.0.0.
-
property
coefficientStandardErrors[source]¶ Standard error of estimated coefficients and intercept.
If
GeneralizedLinearRegression.fitInterceptis set to True, then the last element returned corresponds to the intercept.New in version 2.0.0.
-
property
-
class
pyspark.ml.regression.IsotonicRegression(*args, **kwargs)[source]¶ Currently implemented using parallelized pool adjacent violators algorithm. Only univariate (single feature) algorithm supported.
>>> from pyspark.ml.linalg import Vectors >>> df = spark.createDataFrame([ ... (1.0, Vectors.dense(1.0)), ... (0.0, Vectors.sparse(1, [], []))], ["label", "features"]) >>> ir = IsotonicRegression() >>> model = ir.fit(df) >>> test0 = spark.createDataFrame([(Vectors.dense(-1.0),)], ["features"]) >>> model.transform(test0).head().prediction 0.0 >>> model.boundaries DenseVector([0.0, 1.0]) >>> ir_path = temp_path + "/ir" >>> ir.save(ir_path) >>> ir2 = IsotonicRegression.load(ir_path) >>> ir2.getIsotonic() True >>> model_path = temp_path + "/ir_model" >>> model.save(model_path) >>> model2 = IsotonicRegressionModel.load(model_path) >>> model.boundaries == model2.boundaries True >>> model.predictions == model2.predictions True
New in version 1.6.0.
-
setParams(featuresCol='features', labelCol='label', predictionCol='prediction', weightCol=None, isotonic=True, featureIndex=0)[source]¶ setParams(self, featuresCol=”features”, labelCol=”label”, predictionCol=”prediction”, weightCol=None, isotonic=True, featureIndex=0): Set the params for IsotonicRegression.
-
-
class
pyspark.ml.regression.IsotonicRegressionModel(java_model=None)[source]¶ Model fitted by
IsotonicRegression.New in version 1.6.0.
-
class
pyspark.ml.regression.LinearRegression(*args, **kwargs)[source]¶ Linear regression.
The learning objective is to minimize the specified loss function, with regularization. This supports two kinds of loss:
squaredError (a.k.a squared loss)
huber (a hybrid of squared error for relatively small errors and absolute error for relatively large ones, and we estimate the scale parameter from training data)
This supports multiple types of regularization:
none (a.k.a. ordinary least squares)
L2 (ridge regression)
L1 (Lasso)
L2 + L1 (elastic net)
Note: Fitting with huber loss only supports none and L2 regularization.
>>> from pyspark.ml.linalg import Vectors >>> df = spark.createDataFrame([ ... (1.0, 2.0, Vectors.dense(1.0)), ... (0.0, 2.0, Vectors.sparse(1, [], []))], ["label", "weight", "features"]) >>> lr = LinearRegression(maxIter=5, regParam=0.0, solver="normal", weightCol="weight") >>> model = lr.fit(df) >>> test0 = spark.createDataFrame([(Vectors.dense(-1.0),)], ["features"]) >>> abs(model.transform(test0).head().prediction - (-1.0)) < 0.001 True >>> abs(model.coefficients[0] - 1.0) < 0.001 True >>> abs(model.intercept - 0.0) < 0.001 True >>> test1 = spark.createDataFrame([(Vectors.sparse(1, [0], [1.0]),)], ["features"]) >>> abs(model.transform(test1).head().prediction - 1.0) < 0.001 True >>> lr.setParams("vector") Traceback (most recent call last): ... TypeError: Method setParams forces keyword arguments. >>> lr_path = temp_path + "/lr" >>> lr.save(lr_path) >>> lr2 = LinearRegression.load(lr_path) >>> lr2.getMaxIter() 5 >>> model_path = temp_path + "/lr_model" >>> model.save(model_path) >>> model2 = LinearRegressionModel.load(model_path) >>> model.coefficients[0] == model2.coefficients[0] True >>> model.intercept == model2.intercept True >>> model.numFeatures 1 >>> model.write().format("pmml").save(model_path + "_2")
New in version 1.4.0.
-
setParams(self, featuresCol='features', labelCol='label', predictionCol='prediction', maxIter=100, regParam=0.0, elasticNetParam=0.0, tol=1e-06, fitIntercept=True, standardization=True, solver='auto', weightCol=None, aggregationDepth=2, loss='squaredError', epsilon=1.35)[source]¶ Sets params for linear regression.
New in version 1.4.0.
-
class
pyspark.ml.regression.LinearRegressionModel(java_model=None)[source]¶ Model fitted by
LinearRegression.New in version 1.4.0.
-
evaluate(dataset)[source]¶ Evaluates the model on a test dataset.
- Parameters
dataset – Test dataset to evaluate model on, where dataset is an instance of
pyspark.sql.DataFrame
New in version 2.0.0.
-
property
hasSummary[source]¶ Indicates whether a training summary exists for this model instance.
New in version 2.0.0.
-
-
class
pyspark.ml.regression.LinearRegressionSummary(java_obj=None)[source]¶ Note
Experimental
Linear regression results evaluated on a dataset.
New in version 2.0.0.
-
property
coefficientStandardErrors[source]¶ Standard error of estimated coefficients and intercept. This value is only available when using the “normal” solver.
If
LinearRegression.fitInterceptis set to True, then the last element returned corresponds to the intercept.See also
LinearRegression.solverNew in version 2.0.0.
-
property
devianceResiduals[source]¶ The weighted residuals, the usual residuals rescaled by the square root of the instance weights.
New in version 2.0.0.
-
property
explainedVariance[source]¶ Returns the explained variance regression score. explainedVariance =
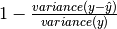
See also
Note
This ignores instance weights (setting all to 1.0) from
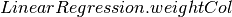 . This will change in later Spark
versions.
. This will change in later Spark
versions.New in version 2.0.0.
-
property
featuresCol[source]¶ Field in “predictions” which gives the features of each instance as a vector.
New in version 2.0.0.
-
property
labelCol[source]¶ Field in “predictions” which gives the true label of each instance.
New in version 2.0.0.
-
property
meanAbsoluteError[source]¶ Returns the mean absolute error, which is a risk function corresponding to the expected value of the absolute error loss or l1-norm loss.
Note
This ignores instance weights (setting all to 1.0) from
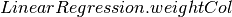 . This will change in later Spark
versions.
. This will change in later Spark
versions.New in version 2.0.0.
-
property
meanSquaredError[source]¶ Returns the mean squared error, which is a risk function corresponding to the expected value of the squared error loss or quadratic loss.
Note
This ignores instance weights (setting all to 1.0) from
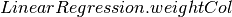 . This will change in later Spark
versions.
. This will change in later Spark
versions.New in version 2.0.0.
-
property
pValues[source]¶ Two-sided p-value of estimated coefficients and intercept. This value is only available when using the “normal” solver.
If
LinearRegression.fitInterceptis set to True, then the last element returned corresponds to the intercept.See also
LinearRegression.solverNew in version 2.0.0.
-
property
predictionCol[source]¶ Field in “predictions” which gives the predicted value of the label at each instance.
New in version 2.0.0.
-
property
r2[source]¶ Returns R^2, the coefficient of determination.
Note
This ignores instance weights (setting all to 1.0) from
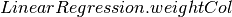 . This will change in later Spark
versions.
. This will change in later Spark
versions.New in version 2.0.0.
-
property
r2adj[source]¶ Returns Adjusted R^2, the adjusted coefficient of determination.
Note
This ignores instance weights (setting all to 1.0) from
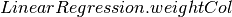 . This will change in later Spark versions.
. This will change in later Spark versions.New in version 2.4.0.
-
property
rootMeanSquaredError[source]¶ Returns the root mean squared error, which is defined as the square root of the mean squared error.
Note
This ignores instance weights (setting all to 1.0) from
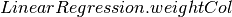 . This will change in later Spark
versions.
. This will change in later Spark
versions.New in version 2.0.0.
-
property
tValues[source]¶ T-statistic of estimated coefficients and intercept. This value is only available when using the “normal” solver.
If
LinearRegression.fitInterceptis set to True, then the last element returned corresponds to the intercept.See also
LinearRegression.solverNew in version 2.0.0.
-
property
-
class
pyspark.ml.regression.LinearRegressionTrainingSummary(java_obj=None)[source]¶ Note
Experimental
Linear regression training results. Currently, the training summary ignores the training weights except for the objective trace.
New in version 2.0.0.
-
class
pyspark.ml.regression.RandomForestRegressionModel(java_model=None)[source]¶ Model fitted by
RandomForestRegressor.New in version 1.4.0.
-
property
featureImportances[source]¶ Estimate of the importance of each feature.
Each feature’s importance is the average of its importance across all trees in the ensemble The importance vector is normalized to sum to 1. This method is suggested by Hastie et al. (Hastie, Tibshirani, Friedman. “The Elements of Statistical Learning, 2nd Edition.” 2001.) and follows the implementation from scikit-learn.
New in version 2.0.0.
-
property
-
class
pyspark.ml.regression.RandomForestRegressor(*args, **kwargs)[source]¶ Random Forest learning algorithm for regression. It supports both continuous and categorical features.
>>> from numpy import allclose >>> from pyspark.ml.linalg import Vectors >>> df = spark.createDataFrame([ ... (1.0, Vectors.dense(1.0)), ... (0.0, Vectors.sparse(1, [], []))], ["label", "features"]) >>> rf = RandomForestRegressor(numTrees=2, maxDepth=2, seed=42) >>> model = rf.fit(df) >>> model.featureImportances SparseVector(1, {0: 1.0}) >>> allclose(model.treeWeights, [1.0, 1.0]) True >>> test0 = spark.createDataFrame([(Vectors.dense(-1.0),)], ["features"]) >>> model.transform(test0).head().prediction 0.0 >>> model.numFeatures 1 >>> model.trees [DecisionTreeRegressionModel (uid=...) of depth..., DecisionTreeRegressionModel...] >>> model.getNumTrees 2 >>> test1 = spark.createDataFrame([(Vectors.sparse(1, [0], [1.0]),)], ["features"]) >>> model.transform(test1).head().prediction 0.5 >>> rfr_path = temp_path + "/rfr" >>> rf.save(rfr_path) >>> rf2 = RandomForestRegressor.load(rfr_path) >>> rf2.getNumTrees() 2 >>> model_path = temp_path + "/rfr_model" >>> model.save(model_path) >>> model2 = RandomForestRegressionModel.load(model_path) >>> model.featureImportances == model2.featureImportances True
New in version 1.4.0.
-
setFeatureSubsetStrategy(value)[source]¶ Sets the value of
featureSubsetStrategy.New in version 2.4.0.
-
setParams(self, featuresCol='features', labelCol='label', predictionCol='prediction', maxDepth=5, maxBins=32, minInstancesPerNode=1, minInfoGain=0.0, maxMemoryInMB=256, cacheNodeIds=False, checkpointInterval=10, impurity='variance', subsamplingRate=1.0, seed=None, numTrees=20, featureSubsetStrategy='auto')[source]¶ Sets params for linear regression.
New in version 1.4.0.
-
27.3. Classification API¶
-
class
pyspark.ml.classification.BinaryLogisticRegressionSummary(java_obj=None)[source]¶ Note
Experimental
Binary Logistic regression results for a given model.
New in version 2.0.0.
-
property
areaUnderROC[source]¶ Computes the area under the receiver operating characteristic (ROC) curve.
Note
This ignores instance weights (setting all to 1.0) from
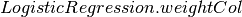 . This will change in later Spark
versions.
. This will change in later Spark
versions.New in version 2.0.0.
-
property
fMeasureByThreshold[source]¶ Returns a dataframe with two fields (threshold, F-Measure) curve with beta = 1.0.
Note
This ignores instance weights (setting all to 1.0) from
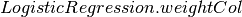 . This will change in later Spark
versions.
. This will change in later Spark
versions.New in version 2.0.0.
-
property
pr[source]¶ Returns the precision-recall curve, which is a Dataframe containing two fields recall, precision with (0.0, 1.0) prepended to it.
Note
This ignores instance weights (setting all to 1.0) from
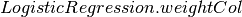 . This will change in later Spark
versions.
. This will change in later Spark
versions.New in version 2.0.0.
-
property
precisionByThreshold[source]¶ Returns a dataframe with two fields (threshold, precision) curve. Every possible probability obtained in transforming the dataset are used as thresholds used in calculating the precision.
Note
This ignores instance weights (setting all to 1.0) from
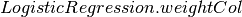 . This will change in later Spark
versions.
. This will change in later Spark
versions.New in version 2.0.0.
-
property
recallByThreshold[source]¶ Returns a dataframe with two fields (threshold, recall) curve. Every possible probability obtained in transforming the dataset are used as thresholds used in calculating the recall.
Note
This ignores instance weights (setting all to 1.0) from
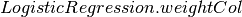 . This will change in later Spark
versions.
. This will change in later Spark
versions.New in version 2.0.0.
-
property
roc[source]¶ Returns the receiver operating characteristic (ROC) curve, which is a Dataframe having two fields (FPR, TPR) with (0.0, 0.0) prepended and (1.0, 1.0) appended to it.
See also
Note
This ignores instance weights (setting all to 1.0) from
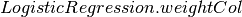 . This will change in later Spark
versions.
. This will change in later Spark
versions.New in version 2.0.0.
-
property
-
class
pyspark.ml.classification.BinaryLogisticRegressionTrainingSummary(java_obj=None)[source]¶ Note
Experimental
Binary Logistic regression training results for a given model.
New in version 2.0.0.
-
class
pyspark.ml.classification.DecisionTreeClassificationModel(java_model=None)[source]¶ Model fitted by DecisionTreeClassifier.
New in version 1.4.0.
-
property
featureImportances[source]¶ Estimate of the importance of each feature.
This generalizes the idea of “Gini” importance to other losses, following the explanation of Gini importance from “Random Forests” documentation by Leo Breiman and Adele Cutler, and following the implementation from scikit-learn.
- This feature importance is calculated as follows:
importance(feature j) = sum (over nodes which split on feature j) of the gain, where gain is scaled by the number of instances passing through node
Normalize importances for tree to sum to 1.
Note
Feature importance for single decision trees can have high variance due to correlated predictor variables. Consider using a
RandomForestClassifierto determine feature importance instead.New in version 2.0.0.
-
property
-
class
pyspark.ml.classification.DecisionTreeClassifier(*args, **kwargs)[source]¶ Decision tree learning algorithm for classification. It supports both binary and multiclass labels, as well as both continuous and categorical features.
>>> from pyspark.ml.linalg import Vectors >>> from pyspark.ml.feature import StringIndexer >>> df = spark.createDataFrame([ ... (1.0, Vectors.dense(1.0)), ... (0.0, Vectors.sparse(1, [], []))], ["label", "features"]) >>> stringIndexer = StringIndexer(inputCol="label", outputCol="indexed") >>> si_model = stringIndexer.fit(df) >>> td = si_model.transform(df) >>> dt = DecisionTreeClassifier(maxDepth=2, labelCol="indexed") >>> model = dt.fit(td) >>> model.numNodes 3 >>> model.depth 1 >>> model.featureImportances SparseVector(1, {0: 1.0}) >>> model.numFeatures 1 >>> model.numClasses 2 >>> print(model.toDebugString) DecisionTreeClassificationModel (uid=...) of depth 1 with 3 nodes... >>> test0 = spark.createDataFrame([(Vectors.dense(-1.0),)], ["features"]) >>> result = model.transform(test0).head() >>> result.prediction 0.0 >>> result.probability DenseVector([1.0, 0.0]) >>> result.rawPrediction DenseVector([1.0, 0.0]) >>> test1 = spark.createDataFrame([(Vectors.sparse(1, [0], [1.0]),)], ["features"]) >>> model.transform(test1).head().prediction 1.0
>>> dtc_path = temp_path + "/dtc" >>> dt.save(dtc_path) >>> dt2 = DecisionTreeClassifier.load(dtc_path) >>> dt2.getMaxDepth() 2 >>> model_path = temp_path + "/dtc_model" >>> model.save(model_path) >>> model2 = DecisionTreeClassificationModel.load(model_path) >>> model.featureImportances == model2.featureImportances True
New in version 1.4.0.
-
setParams(self, featuresCol='features', labelCol='label', predictionCol='prediction', probabilityCol='probability', rawPredictionCol='rawPrediction', maxDepth=5, maxBins=32, minInstancesPerNode=1, minInfoGain=0.0, maxMemoryInMB=256, cacheNodeIds=False, checkpointInterval=10, impurity='gini', seed=None)[source]¶ Sets params for the DecisionTreeClassifier.
New in version 1.4.0.
-
-
class
pyspark.ml.classification.GBTClassificationModel(java_model=None)[source]¶ Model fitted by GBTClassifier.
New in version 1.4.0.
-
evaluateEachIteration(dataset)[source]¶ Method to compute error or loss for every iteration of gradient boosting.
- Parameters
dataset – Test dataset to evaluate model on, where dataset is an instance of
pyspark.sql.DataFrame
New in version 2.4.0.
-
property
featureImportances[source]¶ Estimate of the importance of each feature.
Each feature’s importance is the average of its importance across all trees in the ensemble The importance vector is normalized to sum to 1. This method is suggested by Hastie et al. (Hastie, Tibshirani, Friedman. “The Elements of Statistical Learning, 2nd Edition.” 2001.) and follows the implementation from scikit-learn.
New in version 2.0.0.
-
-
class
pyspark.ml.classification.GBTClassifier(*args, **kwargs)[source]¶ Gradient-Boosted Trees (GBTs) learning algorithm for classification. It supports binary labels, as well as both continuous and categorical features.
The implementation is based upon: J.H. Friedman. “Stochastic Gradient Boosting.” 1999.
Notes on Gradient Boosting vs. TreeBoost: - This implementation is for Stochastic Gradient Boosting, not for TreeBoost. - Both algorithms learn tree ensembles by minimizing loss functions. - TreeBoost (Friedman, 1999) additionally modifies the outputs at tree leaf nodes based on the loss function, whereas the original gradient boosting method does not. - We expect to implement TreeBoost in the future: SPARK-4240
Note
Multiclass labels are not currently supported.
>>> from numpy import allclose >>> from pyspark.ml.linalg import Vectors >>> from pyspark.ml.feature import StringIndexer >>> df = spark.createDataFrame([ ... (1.0, Vectors.dense(1.0)), ... (0.0, Vectors.sparse(1, [], []))], ["label", "features"]) >>> stringIndexer = StringIndexer(inputCol="label", outputCol="indexed") >>> si_model = stringIndexer.fit(df) >>> td = si_model.transform(df) >>> gbt = GBTClassifier(maxIter=5, maxDepth=2, labelCol="indexed", seed=42) >>> gbt.getFeatureSubsetStrategy() 'all' >>> model = gbt.fit(td) >>> model.featureImportances SparseVector(1, {0: 1.0}) >>> allclose(model.treeWeights, [1.0, 0.1, 0.1, 0.1, 0.1]) True >>> test0 = spark.createDataFrame([(Vectors.dense(-1.0),)], ["features"]) >>> model.transform(test0).head().prediction 0.0 >>> test1 = spark.createDataFrame([(Vectors.sparse(1, [0], [1.0]),)], ["features"]) >>> model.transform(test1).head().prediction 1.0 >>> model.totalNumNodes 15 >>> print(model.toDebugString) GBTClassificationModel (uid=...)...with 5 trees... >>> gbtc_path = temp_path + "gbtc" >>> gbt.save(gbtc_path) >>> gbt2 = GBTClassifier.load(gbtc_path) >>> gbt2.getMaxDepth() 2 >>> model_path = temp_path + "gbtc_model" >>> model.save(model_path) >>> model2 = GBTClassificationModel.load(model_path) >>> model.featureImportances == model2.featureImportances True >>> model.treeWeights == model2.treeWeights True >>> model.trees [DecisionTreeRegressionModel (uid=...) of depth..., DecisionTreeRegressionModel...] >>> validation = spark.createDataFrame([(0.0, Vectors.dense(-1.0),)], ... ["indexed", "features"]) >>> model.evaluateEachIteration(validation) [0.25..., 0.23..., 0.21..., 0.19..., 0.18...] >>> model.numClasses 2
New in version 1.4.0.
-
setFeatureSubsetStrategy(value)[source]¶ Sets the value of
featureSubsetStrategy.New in version 2.4.0.
-
setParams(self, featuresCol='features', labelCol='label', predictionCol='prediction', maxDepth=5, maxBins=32, minInstancesPerNode=1, minInfoGain=0.0, maxMemoryInMB=256, cacheNodeIds=False, checkpointInterval=10, lossType='logistic', maxIter=20, stepSize=0.1, seed=None, subsamplingRate=1.0, featureSubsetStrategy='all')[source]¶ Sets params for Gradient Boosted Tree Classification.
New in version 1.4.0.
-
-
class
pyspark.ml.classification.LinearSVC(*args, **kwargs)[source]¶ Note
Experimental
This binary classifier optimizes the Hinge Loss using the OWLQN optimizer. Only supports L2 regularization currently.
>>> from pyspark.sql import Row >>> from pyspark.ml.linalg import Vectors >>> df = sc.parallelize([ ... Row(label=1.0, features=Vectors.dense(1.0, 1.0, 1.0)), ... Row(label=0.0, features=Vectors.dense(1.0, 2.0, 3.0))]).toDF() >>> svm = LinearSVC(maxIter=5, regParam=0.01) >>> model = svm.fit(df) >>> model.coefficients DenseVector([0.0, -0.2792, -0.1833]) >>> model.intercept 1.0206118982229047 >>> model.numClasses 2 >>> model.numFeatures 3 >>> test0 = sc.parallelize([Row(features=Vectors.dense(-1.0, -1.0, -1.0))]).toDF() >>> result = model.transform(test0).head() >>> result.prediction 1.0 >>> result.rawPrediction DenseVector([-1.4831, 1.4831]) >>> svm_path = temp_path + "/svm" >>> svm.save(svm_path) >>> svm2 = LinearSVC.load(svm_path) >>> svm2.getMaxIter() 5 >>> model_path = temp_path + "/svm_model" >>> model.save(model_path) >>> model2 = LinearSVCModel.load(model_path) >>> model.coefficients[0] == model2.coefficients[0] True >>> model.intercept == model2.intercept True
New in version 2.2.0.
-
setParams(featuresCol='features', labelCol='label', predictionCol='prediction', maxIter=100, regParam=0.0, tol=1e-06, rawPredictionCol='rawPrediction', fitIntercept=True, standardization=True, threshold=0.0, weightCol=None, aggregationDepth=2)[source]¶ setParams(self, featuresCol=”features”, labelCol=”label”, predictionCol=”prediction”, maxIter=100, regParam=0.0, tol=1e-6, rawPredictionCol=”rawPrediction”, fitIntercept=True, standardization=True, threshold=0.0, weightCol=None, aggregationDepth=2): Sets params for Linear SVM Classifier.
New in version 2.2.0.
-
-
class
pyspark.ml.classification.LinearSVCModel(java_model=None)[source]¶ Note
Experimental
Model fitted by LinearSVC.
New in version 2.2.0.
-
class
pyspark.ml.classification.LogisticRegression(*args, **kwargs)[source]¶ Logistic regression. This class supports multinomial logistic (softmax) and binomial logistic regression.
>>> from pyspark.sql import Row >>> from pyspark.ml.linalg import Vectors >>> bdf = sc.parallelize([ ... Row(label=1.0, weight=1.0, features=Vectors.dense(0.0, 5.0)), ... Row(label=0.0, weight=2.0, features=Vectors.dense(1.0, 2.0)), ... Row(label=1.0, weight=3.0, features=Vectors.dense(2.0, 1.0)), ... Row(label=0.0, weight=4.0, features=Vectors.dense(3.0, 3.0))]).toDF() >>> blor = LogisticRegression(regParam=0.01, weightCol="weight") >>> blorModel = blor.fit(bdf) >>> blorModel.coefficients DenseVector([-1.080..., -0.646...]) >>> blorModel.intercept 3.112... >>> data_path = "data/mllib/sample_multiclass_classification_data.txt" >>> mdf = spark.read.format("libsvm").load(data_path) >>> mlor = LogisticRegression(regParam=0.1, elasticNetParam=1.0, family="multinomial") >>> mlorModel = mlor.fit(mdf) >>> mlorModel.coefficientMatrix SparseMatrix(3, 4, [0, 1, 2, 3], [3, 2, 1], [1.87..., -2.75..., -0.50...], 1) >>> mlorModel.interceptVector DenseVector([0.04..., -0.42..., 0.37...]) >>> test0 = sc.parallelize([Row(features=Vectors.dense(-1.0, 1.0))]).toDF() >>> result = blorModel.transform(test0).head() >>> result.prediction 1.0 >>> result.probability DenseVector([0.02..., 0.97...]) >>> result.rawPrediction DenseVector([-3.54..., 3.54...]) >>> test1 = sc.parallelize([Row(features=Vectors.sparse(2, [0], [1.0]))]).toDF() >>> blorModel.transform(test1).head().prediction 1.0 >>> blor.setParams("vector") Traceback (most recent call last): ... TypeError: Method setParams forces keyword arguments. >>> lr_path = temp_path + "/lr" >>> blor.save(lr_path) >>> lr2 = LogisticRegression.load(lr_path) >>> lr2.getRegParam() 0.01 >>> model_path = temp_path + "/lr_model" >>> blorModel.save(model_path) >>> model2 = LogisticRegressionModel.load(model_path) >>> blorModel.coefficients[0] == model2.coefficients[0] True >>> blorModel.intercept == model2.intercept True >>> model2 LogisticRegressionModel: uid = ..., numClasses = 2, numFeatures = 2
New in version 1.3.0.
-
getLowerBoundsOnCoefficients()[source]¶ Gets the value of
lowerBoundsOnCoefficientsNew in version 2.3.0.
-
getLowerBoundsOnIntercepts()[source]¶ Gets the value of
lowerBoundsOnInterceptsNew in version 2.3.0.
-
getThreshold()[source]¶ Get threshold for binary classification.
If
thresholdsis set with length 2 (i.e., binary classification), this returns the equivalent threshold: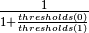 .
Otherwise, returns
.
Otherwise, returns thresholdif set or its default value if unset.New in version 1.4.0.
-
getThresholds()[source]¶ If
thresholdsis set, return its value. Otherwise, ifthresholdis set, return the equivalent thresholds for binary classification: (1-threshold, threshold). If neither are set, throw an error.New in version 1.5.0.
-
getUpperBoundsOnCoefficients()[source]¶ Gets the value of
upperBoundsOnCoefficientsNew in version 2.3.0.
-
getUpperBoundsOnIntercepts()[source]¶ Gets the value of
upperBoundsOnInterceptsNew in version 2.3.0.
-
setLowerBoundsOnCoefficients(value)[source]¶ Sets the value of
lowerBoundsOnCoefficientsNew in version 2.3.0.
-
setLowerBoundsOnIntercepts(value)[source]¶ Sets the value of
lowerBoundsOnInterceptsNew in version 2.3.0.
-
setParams(featuresCol='features', labelCol='label', predictionCol='prediction', maxIter=100, regParam=0.0, elasticNetParam=0.0, tol=1e-06, fitIntercept=True, threshold=0.5, thresholds=None, probabilityCol='probability', rawPredictionCol='rawPrediction', standardization=True, weightCol=None, aggregationDepth=2, family='auto', lowerBoundsOnCoefficients=None, upperBoundsOnCoefficients=None, lowerBoundsOnIntercepts=None, upperBoundsOnIntercepts=None)[source]¶ setParams(self, featuresCol=”features”, labelCol=”label”, predictionCol=”prediction”, maxIter=100, regParam=0.0, elasticNetParam=0.0, tol=1e-6, fitIntercept=True, threshold=0.5, thresholds=None, probabilityCol=”probability”, rawPredictionCol=”rawPrediction”, standardization=True, weightCol=None, aggregationDepth=2, family=”auto”, lowerBoundsOnCoefficients=None, upperBoundsOnCoefficients=None, lowerBoundsOnIntercepts=None, upperBoundsOnIntercepts=None): Sets params for logistic regression. If the threshold and thresholds Params are both set, they must be equivalent.
New in version 1.3.0.
-
setThreshold(value)[source]¶ Sets the value of
threshold. Clears value ofthresholdsif it has been set.New in version 1.4.0.
-
setThresholds(value)[source]¶ Sets the value of
thresholds. Clears value ofthresholdif it has been set.New in version 1.5.0.
-
-
class
pyspark.ml.classification.LogisticRegressionModel(java_model=None)[source]¶ Model fitted by LogisticRegression.
New in version 1.3.0.
-
property
coefficients[source]¶ Model coefficients of binomial logistic regression. An exception is thrown in the case of multinomial logistic regression.
New in version 2.0.0.
-
evaluate(dataset)[source]¶ Evaluates the model on a test dataset.
- Parameters
dataset – Test dataset to evaluate model on, where dataset is an instance of
pyspark.sql.DataFrame
New in version 2.0.0.
-
property
hasSummary[source]¶ Indicates whether a training summary exists for this model instance.
New in version 2.0.0.
-
property
-
class
pyspark.ml.classification.LogisticRegressionSummary(java_obj=None)[source]¶ Note
Experimental
Abstraction for Logistic Regression Results for a given model.
New in version 2.0.0.
-
property
accuracy[source]¶ Returns accuracy. (equals to the total number of correctly classified instances out of the total number of instances.)
New in version 2.3.0.
-
fMeasureByLabel(beta=1.0)[source]¶ Returns f-measure for each label (category).
New in version 2.3.0.
-
property
falsePositiveRateByLabel[source]¶ Returns false positive rate for each label (category).
New in version 2.3.0.
-
property
featuresCol[source]¶ Field in “predictions” which gives the features of each instance as a vector.
New in version 2.0.0.
-
property
labelCol[source]¶ Field in “predictions” which gives the true label of each instance.
New in version 2.0.0.
-
property
labels[source]¶ Returns the sequence of labels in ascending order. This order matches the order used in metrics which are specified as arrays over labels, e.g., truePositiveRateByLabel.
Note: In most cases, it will be values {0.0, 1.0, …, numClasses-1}, However, if the training set is missing a label, then all of the arrays over labels (e.g., from truePositiveRateByLabel) will be of length numClasses-1 instead of the expected numClasses.
New in version 2.3.0.
-
property
precisionByLabel[source]¶ Returns precision for each label (category).
New in version 2.3.0.
-
property
predictionCol[source]¶ Field in “predictions” which gives the prediction of each class.
New in version 2.3.0.
-
property
probabilityCol[source]¶ Field in “predictions” which gives the probability of each class as a vector.
New in version 2.0.0.
-
property
truePositiveRateByLabel[source]¶ Returns true positive rate for each label (category).
New in version 2.3.0.
-
property
weightedFalsePositiveRate[source]¶ Returns weighted false positive rate.
New in version 2.3.0.
-
property
-
class
pyspark.ml.classification.LogisticRegressionTrainingSummary(java_obj=None)[source]¶ Note
Experimental
Abstraction for multinomial Logistic Regression Training results. Currently, the training summary ignores the training weights except for the objective trace.
New in version 2.0.0.
-
class
pyspark.ml.classification.MultilayerPerceptronClassificationModel(java_model=None)[source]¶ Model fitted by MultilayerPerceptronClassifier.
New in version 1.6.0.
-
class
pyspark.ml.classification.MultilayerPerceptronClassifier(*args, **kwargs)[source]¶ Classifier trainer based on the Multilayer Perceptron. Each layer has sigmoid activation function, output layer has softmax. Number of inputs has to be equal to the size of feature vectors. Number of outputs has to be equal to the total number of labels.
>>> from pyspark.ml.linalg import Vectors >>> df = spark.createDataFrame([ ... (0.0, Vectors.dense([0.0, 0.0])), ... (1.0, Vectors.dense([0.0, 1.0])), ... (1.0, Vectors.dense([1.0, 0.0])), ... (0.0, Vectors.dense([1.0, 1.0]))], ["label", "features"]) >>> mlp = MultilayerPerceptronClassifier(maxIter=100, layers=[2, 2, 2], blockSize=1, seed=123) >>> model = mlp.fit(df) >>> model.layers [2, 2, 2] >>> model.weights.size 12 >>> testDF = spark.createDataFrame([ ... (Vectors.dense([1.0, 0.0]),), ... (Vectors.dense([0.0, 0.0]),)], ["features"]) >>> model.transform(testDF).select("features", "prediction").show() +---------+----------+ | features|prediction| +---------+----------+ |[1.0,0.0]| 1.0| |[0.0,0.0]| 0.0| +---------+----------+ ... >>> mlp_path = temp_path + "/mlp" >>> mlp.save(mlp_path) >>> mlp2 = MultilayerPerceptronClassifier.load(mlp_path) >>> mlp2.getBlockSize() 1 >>> model_path = temp_path + "/mlp_model" >>> model.save(model_path) >>> model2 = MultilayerPerceptronClassificationModel.load(model_path) >>> model.layers == model2.layers True >>> model.weights == model2.weights True >>> mlp2 = mlp2.setInitialWeights(list(range(0, 12))) >>> model3 = mlp2.fit(df) >>> model3.weights != model2.weights True >>> model3.layers == model.layers True
New in version 1.6.0.
-
getInitialWeights()[source]¶ Gets the value of initialWeights or its default value.
New in version 2.0.0.
-
setParams(featuresCol='features', labelCol='label', predictionCol='prediction', maxIter=100, tol=1e-06, seed=None, layers=None, blockSize=128, stepSize=0.03, solver='l-bfgs', initialWeights=None, probabilityCol='probability', rawPredictionCol='rawPrediction')[source]¶ setParams(self, featuresCol=”features”, labelCol=”label”, predictionCol=”prediction”, maxIter=100, tol=1e-6, seed=None, layers=None, blockSize=128, stepSize=0.03, solver=”l-bfgs”, initialWeights=None, probabilityCol=”probability”, rawPredictionCol=”rawPrediction”): Sets params for MultilayerPerceptronClassifier.
New in version 1.6.0.
-
-
class
pyspark.ml.classification.NaiveBayes(*args, **kwargs)[source]¶ Naive Bayes Classifiers. It supports both Multinomial and Bernoulli NB. Multinomial NB can handle finitely supported discrete data. For example, by converting documents into TF-IDF vectors, it can be used for document classification. By making every vector a binary (0/1) data, it can also be used as Bernoulli NB. The input feature values must be nonnegative.
>>> from pyspark.sql import Row >>> from pyspark.ml.linalg import Vectors >>> df = spark.createDataFrame([ ... Row(label=0.0, weight=0.1, features=Vectors.dense([0.0, 0.0])), ... Row(label=0.0, weight=0.5, features=Vectors.dense([0.0, 1.0])), ... Row(label=1.0, weight=1.0, features=Vectors.dense([1.0, 0.0]))]) >>> nb = NaiveBayes(smoothing=1.0, modelType="multinomial", weightCol="weight") >>> model = nb.fit(df) >>> model.pi DenseVector([-0.81..., -0.58...]) >>> model.theta DenseMatrix(2, 2, [-0.91..., -0.51..., -0.40..., -1.09...], 1) >>> test0 = sc.parallelize([Row(features=Vectors.dense([1.0, 0.0]))]).toDF() >>> result = model.transform(test0).head() >>> result.prediction 1.0 >>> result.probability DenseVector([0.32..., 0.67...]) >>> result.rawPrediction DenseVector([-1.72..., -0.99...]) >>> test1 = sc.parallelize([Row(features=Vectors.sparse(2, [0], [1.0]))]).toDF() >>> model.transform(test1).head().prediction 1.0 >>> nb_path = temp_path + "/nb" >>> nb.save(nb_path) >>> nb2 = NaiveBayes.load(nb_path) >>> nb2.getSmoothing() 1.0 >>> model_path = temp_path + "/nb_model" >>> model.save(model_path) >>> model2 = NaiveBayesModel.load(model_path) >>> model.pi == model2.pi True >>> model.theta == model2.theta True >>> nb = nb.setThresholds([0.01, 10.00]) >>> model3 = nb.fit(df) >>> result = model3.transform(test0).head() >>> result.prediction 0.0
New in version 1.5.0.
-
class
pyspark.ml.classification.NaiveBayesModel(java_model=None)[source]¶ Model fitted by NaiveBayes.
New in version 1.5.0.
-
class
pyspark.ml.classification.OneVsRest(*args, **kwargs)[source]¶ Note
Experimental
Reduction of Multiclass Classification to Binary Classification. Performs reduction using one against all strategy. For a multiclass classification with k classes, train k models (one per class). Each example is scored against all k models and the model with highest score is picked to label the example.
>>> from pyspark.sql import Row >>> from pyspark.ml.linalg import Vectors >>> data_path = "data/mllib/sample_multiclass_classification_data.txt" >>> df = spark.read.format("libsvm").load(data_path) >>> lr = LogisticRegression(regParam=0.01) >>> ovr = OneVsRest(classifier=lr) >>> model = ovr.fit(df) >>> model.models[0].coefficients DenseVector([0.5..., -1.0..., 3.4..., 4.2...]) >>> model.models[1].coefficients DenseVector([-2.1..., 3.1..., -2.6..., -2.3...]) >>> model.models[2].coefficients DenseVector([0.3..., -3.4..., 1.0..., -1.1...]) >>> [x.intercept for x in model.models] [-2.7..., -2.5..., -1.3...] >>> test0 = sc.parallelize([Row(features=Vectors.dense(-1.0, 0.0, 1.0, 1.0))]).toDF() >>> model.transform(test0).head().prediction 0.0 >>> test1 = sc.parallelize([Row(features=Vectors.sparse(4, [0], [1.0]))]).toDF() >>> model.transform(test1).head().prediction 2.0 >>> test2 = sc.parallelize([Row(features=Vectors.dense(0.5, 0.4, 0.3, 0.2))]).toDF() >>> model.transform(test2).head().prediction 0.0 >>> model_path = temp_path + "/ovr_model" >>> model.save(model_path) >>> model2 = OneVsRestModel.load(model_path) >>> model2.transform(test0).head().prediction 0.0
New in version 2.0.0.
-
copy(extra=None)[source]¶ Creates a copy of this instance with a randomly generated uid and some extra params. This creates a deep copy of the embedded paramMap, and copies the embedded and extra parameters over.
- Parameters
extra – Extra parameters to copy to the new instance
- Returns
Copy of this instance
New in version 2.0.0.
-
setParams(featuresCol='features', labelCol='label', predictionCol='prediction', classifier=None, weightCol=None, parallelism=1)[source]¶ setParams(self, featuresCol=”features”, labelCol=”label”, predictionCol=”prediction”, classifier=None, weightCol=None, parallelism=1): Sets params for OneVsRest.
New in version 2.0.0.
-
-
class
pyspark.ml.classification.OneVsRestModel(models)[source]¶ Note
Experimental
Model fitted by OneVsRest. This stores the models resulting from training k binary classifiers: one for each class. Each example is scored against all k models, and the model with the highest score is picked to label the example.
New in version 2.0.0.
-
copy(extra=None)[source]¶ Creates a copy of this instance with a randomly generated uid and some extra params. This creates a deep copy of the embedded paramMap, and copies the embedded and extra parameters over.
- Parameters
extra – Extra parameters to copy to the new instance
- Returns
Copy of this instance
New in version 2.0.0.
-
-
class
pyspark.ml.classification.RandomForestClassificationModel(java_model=None)[source]¶ Model fitted by RandomForestClassifier.
New in version 1.4.0.
-
property
featureImportances[source]¶ Estimate of the importance of each feature.
Each feature’s importance is the average of its importance across all trees in the ensemble The importance vector is normalized to sum to 1. This method is suggested by Hastie et al. (Hastie, Tibshirani, Friedman. “The Elements of Statistical Learning, 2nd Edition.” 2001.) and follows the implementation from scikit-learn.
New in version 2.0.0.
-
property
-
class
pyspark.ml.classification.RandomForestClassifier(*args, **kwargs)[source]¶ Random Forest learning algorithm for classification. It supports both binary and multiclass labels, as well as both continuous and categorical features.
>>> import numpy >>> from numpy import allclose >>> from pyspark.ml.linalg import Vectors >>> from pyspark.ml.feature import StringIndexer >>> df = spark.createDataFrame([ ... (1.0, Vectors.dense(1.0)), ... (0.0, Vectors.sparse(1, [], []))], ["label", "features"]) >>> stringIndexer = StringIndexer(inputCol="label", outputCol="indexed") >>> si_model = stringIndexer.fit(df) >>> td = si_model.transform(df) >>> rf = RandomForestClassifier(numTrees=3, maxDepth=2, labelCol="indexed", seed=42) >>> model = rf.fit(td) >>> model.featureImportances SparseVector(1, {0: 1.0}) >>> allclose(model.treeWeights, [1.0, 1.0, 1.0]) True >>> test0 = spark.createDataFrame([(Vectors.dense(-1.0),)], ["features"]) >>> result = model.transform(test0).head() >>> result.prediction 0.0 >>> numpy.argmax(result.probability) 0 >>> numpy.argmax(result.rawPrediction) 0 >>> test1 = spark.createDataFrame([(Vectors.sparse(1, [0], [1.0]),)], ["features"]) >>> model.transform(test1).head().prediction 1.0 >>> model.trees [DecisionTreeClassificationModel (uid=...) of depth..., DecisionTreeClassificationModel...] >>> rfc_path = temp_path + "/rfc" >>> rf.save(rfc_path) >>> rf2 = RandomForestClassifier.load(rfc_path) >>> rf2.getNumTrees() 3 >>> model_path = temp_path + "/rfc_model" >>> model.save(model_path) >>> model2 = RandomForestClassificationModel.load(model_path) >>> model.featureImportances == model2.featureImportances True
New in version 1.4.0.
-
setFeatureSubsetStrategy(value)[source]¶ Sets the value of
featureSubsetStrategy.New in version 2.4.0.
-
setParams(self, featuresCol='features', labelCol='label', predictionCol='prediction', probabilityCol='probability', rawPredictionCol='rawPrediction', maxDepth=5, maxBins=32, minInstancesPerNode=1, minInfoGain=0.0, maxMemoryInMB=256, cacheNodeIds=False, checkpointInterval=10, seed=None, impurity='gini', numTrees=20, featureSubsetStrategy='auto', subsamplingRate=1.0)[source]¶ Sets params for linear classification.
New in version 1.4.0.
-
27.4. Clustering API¶
-
class
pyspark.ml.clustering.BisectingKMeans(*args, **kwargs)[source]¶ A bisecting k-means algorithm based on the paper “A comparison of document clustering techniques” by Steinbach, Karypis, and Kumar, with modification to fit Spark. The algorithm starts from a single cluster that contains all points. Iteratively it finds divisible clusters on the bottom level and bisects each of them using k-means, until there are
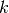 leaf clusters in total or no leaf clusters are divisible.
The bisecting steps of clusters on the same level are grouped together to increase parallelism.
If bisecting all divisible clusters on the bottom level would result more than
leaf clusters in total or no leaf clusters are divisible.
The bisecting steps of clusters on the same level are grouped together to increase parallelism.
If bisecting all divisible clusters on the bottom level would result more than 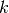 leaf
clusters, larger clusters get higher priority.
leaf
clusters, larger clusters get higher priority.>>> from pyspark.ml.linalg import Vectors >>> data = [(Vectors.dense([0.0, 0.0]),), (Vectors.dense([1.0, 1.0]),), ... (Vectors.dense([9.0, 8.0]),), (Vectors.dense([8.0, 9.0]),)] >>> df = spark.createDataFrame(data, ["features"]) >>> bkm = BisectingKMeans(k=2, minDivisibleClusterSize=1.0) >>> model = bkm.fit(df) >>> centers = model.clusterCenters() >>> len(centers) 2 >>> model.computeCost(df) 2.000... >>> model.hasSummary True >>> summary = model.summary >>> summary.k 2 >>> summary.clusterSizes [2, 2] >>> transformed = model.transform(df).select("features", "prediction") >>> rows = transformed.collect() >>> rows[0].prediction == rows[1].prediction True >>> rows[2].prediction == rows[3].prediction True >>> bkm_path = temp_path + "/bkm" >>> bkm.save(bkm_path) >>> bkm2 = BisectingKMeans.load(bkm_path) >>> bkm2.getK() 2 >>> bkm2.getDistanceMeasure() 'euclidean' >>> model_path = temp_path + "/bkm_model" >>> model.save(model_path) >>> model2 = BisectingKMeansModel.load(model_path) >>> model2.hasSummary False >>> model.clusterCenters()[0] == model2.clusterCenters()[0] array([ True, True], dtype=bool) >>> model.clusterCenters()[1] == model2.clusterCenters()[1] array([ True, True], dtype=bool)
New in version 2.0.0.
-
class
pyspark.ml.clustering.BisectingKMeansModel(java_model=None)[source]¶ Model fitted by BisectingKMeans.
New in version 2.0.0.
-
clusterCenters()[source]¶ Get the cluster centers, represented as a list of NumPy arrays.
New in version 2.0.0.
-
computeCost(dataset)[source]¶ Computes the sum of squared distances between the input points and their corresponding cluster centers.
New in version 2.0.0.
-
-
class
pyspark.ml.clustering.BisectingKMeansSummary(java_obj=None)[source]¶ Note
Experimental
Bisecting KMeans clustering results for a given model.
New in version 2.1.0.
-
class
pyspark.ml.clustering.DistributedLDAModel(java_model=None)[source]¶ Distributed model fitted by
LDA. This type of model is currently only produced by Expectation-Maximization (EM).This model stores the inferred topics, the full training dataset, and the topic distribution for each training document.
New in version 2.0.0.
-
getCheckpointFiles()[source]¶ If using checkpointing and
LDA.keepLastCheckpointis set to true, then there may be saved checkpoint files. This method is provided so that users can manage those files.Note
Removing the checkpoints can cause failures if a partition is lost and is needed by certain
DistributedLDAModelmethods. Reference counting will clean up the checkpoints when this model and derivative data go out of scope.:return List of checkpoint files from training
New in version 2.0.0.
-
logPrior()[source]¶ Log probability of the current parameter estimate: log P(topics, topic distributions for docs | alpha, eta)
New in version 2.0.0.
-
toLocal()[source]¶ Convert this distributed model to a local representation. This discards info about the training dataset.
WARNING: This involves collecting a large
topicsMatrix()to the driver.New in version 2.0.0.
-
trainingLogLikelihood()[source]¶ Log likelihood of the observed tokens in the training set, given the current parameter estimates: log P(docs | topics, topic distributions for docs, Dirichlet hyperparameters)
- Notes:
This excludes the prior; for that, use
logPrior().Even with
logPrior(), this is NOT the same as the data log likelihood given the hyperparameters.This is computed from the topic distributions computed during training. If you call
logLikelihood()on the same training dataset, the topic distributions will be computed again, possibly giving different results.
New in version 2.0.0.
-
-
class
pyspark.ml.clustering.GaussianMixture(*args, **kwargs)[source]¶ GaussianMixture clustering. This class performs expectation maximization for multivariate Gaussian Mixture Models (GMMs). A GMM represents a composite distribution of independent Gaussian distributions with associated “mixing” weights specifying each’s contribution to the composite.
Given a set of sample points, this class will maximize the log-likelihood for a mixture of k Gaussians, iterating until the log-likelihood changes by less than convergenceTol, or until it has reached the max number of iterations. While this process is generally guaranteed to converge, it is not guaranteed to find a global optimum.
Note
For high-dimensional data (with many features), this algorithm may perform poorly. This is due to high-dimensional data (a) making it difficult to cluster at all (based on statistical/theoretical arguments) and (b) numerical issues with Gaussian distributions.
>>> from pyspark.ml.linalg import Vectors
>>> data = [(Vectors.dense([-0.1, -0.05 ]),), ... (Vectors.dense([-0.01, -0.1]),), ... (Vectors.dense([0.9, 0.8]),), ... (Vectors.dense([0.75, 0.935]),), ... (Vectors.dense([-0.83, -0.68]),), ... (Vectors.dense([-0.91, -0.76]),)] >>> df = spark.createDataFrame(data, ["features"]) >>> gm = GaussianMixture(k=3, tol=0.0001, ... maxIter=10, seed=10) >>> model = gm.fit(df) >>> model.hasSummary True >>> summary = model.summary >>> summary.k 3 >>> summary.clusterSizes [2, 2, 2] >>> summary.logLikelihood 8.14636... >>> weights = model.weights >>> len(weights) 3 >>> model.gaussiansDF.select("mean").head() Row(mean=DenseVector([0.825, 0.8675])) >>> model.gaussiansDF.select("cov").head() Row(cov=DenseMatrix(2, 2, [0.0056, -0.0051, -0.0051, 0.0046], False)) >>> transformed = model.transform(df).select("features", "prediction") >>> rows = transformed.collect() >>> rows[4].prediction == rows[5].prediction True >>> rows[2].prediction == rows[3].prediction True >>> gmm_path = temp_path + "/gmm" >>> gm.save(gmm_path) >>> gm2 = GaussianMixture.load(gmm_path) >>> gm2.getK() 3 >>> model_path = temp_path + "/gmm_model" >>> model.save(model_path) >>> model2 = GaussianMixtureModel.load(model_path) >>> model2.hasSummary False >>> model2.weights == model.weights True >>> model2.gaussiansDF.select("mean").head() Row(mean=DenseVector([0.825, 0.8675])) >>> model2.gaussiansDF.select("cov").head() Row(cov=DenseMatrix(2, 2, [0.0056, -0.0051, -0.0051, 0.0046], False))
New in version 2.0.0.
-
class
pyspark.ml.clustering.GaussianMixtureModel(java_model=None)[source]¶ Model fitted by GaussianMixture.
New in version 2.0.0.
-
property
gaussiansDF[source]¶ Retrieve Gaussian distributions as a DataFrame. Each row represents a Gaussian Distribution. The DataFrame has two columns: mean (Vector) and cov (Matrix).
New in version 2.0.0.
-
property
hasSummary[source]¶ Indicates whether a training summary exists for this model instance.
New in version 2.1.0.
-
property
-
class
pyspark.ml.clustering.GaussianMixtureSummary(java_obj=None)[source]¶ Note
Experimental
Gaussian mixture clustering results for a given model.
New in version 2.1.0.
-
property
logLikelihood[source]¶ Total log-likelihood for this model on the given data.
New in version 2.2.0.
-
property
-
class
pyspark.ml.clustering.KMeans(*args, **kwargs)[source]¶ K-means clustering with a k-means++ like initialization mode (the k-means|| algorithm by Bahmani et al).
>>> from pyspark.ml.linalg import Vectors >>> data = [(Vectors.dense([0.0, 0.0]),), (Vectors.dense([1.0, 1.0]),), ... (Vectors.dense([9.0, 8.0]),), (Vectors.dense([8.0, 9.0]),)] >>> df = spark.createDataFrame(data, ["features"]) >>> kmeans = KMeans(k=2, seed=1) >>> model = kmeans.fit(df) >>> centers = model.clusterCenters() >>> len(centers) 2 >>> model.computeCost(df) 2.000... >>> transformed = model.transform(df).select("features", "prediction") >>> rows = transformed.collect() >>> rows[0].prediction == rows[1].prediction True >>> rows[2].prediction == rows[3].prediction True >>> model.hasSummary True >>> summary = model.summary >>> summary.k 2 >>> summary.clusterSizes [2, 2] >>> summary.trainingCost 2.000... >>> kmeans_path = temp_path + "/kmeans" >>> kmeans.save(kmeans_path) >>> kmeans2 = KMeans.load(kmeans_path) >>> kmeans2.getK() 2 >>> model_path = temp_path + "/kmeans_model" >>> model.save(model_path) >>> model2 = KMeansModel.load(model_path) >>> model2.hasSummary False >>> model.clusterCenters()[0] == model2.clusterCenters()[0] array([ True, True], dtype=bool) >>> model.clusterCenters()[1] == model2.clusterCenters()[1] array([ True, True], dtype=bool)
New in version 1.5.0.
-
class
pyspark.ml.clustering.KMeansModel(java_model=None)[source]¶ Model fitted by KMeans.
New in version 1.5.0.
-
clusterCenters()[source]¶ Get the cluster centers, represented as a list of NumPy arrays.
New in version 1.5.0.
-
computeCost(dataset)[source]¶ Return the K-means cost (sum of squared distances of points to their nearest center) for this model on the given data.
- ..note:: Deprecated in 2.4.0. It will be removed in 3.0.0. Use ClusteringEvaluator instead.
You can also get the cost on the training dataset in the summary.
New in version 2.0.0.
-
-
class
pyspark.ml.clustering.LDA(*args, **kwargs)[source]¶ Latent Dirichlet Allocation (LDA), a topic model designed for text documents.
Terminology:
“term” = “word”: an element of the vocabulary
“token”: instance of a term appearing in a document
“topic”: multinomial distribution over terms representing some concept
“document”: one piece of text, corresponding to one row in the input data
- Original LDA paper (journal version):
Blei, Ng, and Jordan. “Latent Dirichlet Allocation.” JMLR, 2003.
Input data (featuresCol): LDA is given a collection of documents as input data, via the featuresCol parameter. Each document is specified as a
Vectorof length vocabSize, where each entry is the count for the corresponding term (word) in the document. Feature transformers such aspyspark.ml.feature.Tokenizerandpyspark.ml.feature.CountVectorizercan be useful for converting text to word count vectors.>>> from pyspark.ml.linalg import Vectors, SparseVector >>> from pyspark.ml.clustering import LDA >>> df = spark.createDataFrame([[1, Vectors.dense([0.0, 1.0])], ... [2, SparseVector(2, {0: 1.0})],], ["id", "features"]) >>> lda = LDA(k=2, seed=1, optimizer="em") >>> model = lda.fit(df) >>> model.isDistributed() True >>> localModel = model.toLocal() >>> localModel.isDistributed() False >>> model.vocabSize() 2 >>> model.describeTopics().show() +-----+-----------+--------------------+ |topic|termIndices| termWeights| +-----+-----------+--------------------+ | 0| [1, 0]|[0.50401530077160...| | 1| [0, 1]|[0.50401530077160...| +-----+-----------+--------------------+ ... >>> model.topicsMatrix() DenseMatrix(2, 2, [0.496, 0.504, 0.504, 0.496], 0) >>> lda_path = temp_path + "/lda" >>> lda.save(lda_path) >>> sameLDA = LDA.load(lda_path) >>> distributed_model_path = temp_path + "/lda_distributed_model" >>> model.save(distributed_model_path) >>> sameModel = DistributedLDAModel.load(distributed_model_path) >>> local_model_path = temp_path + "/lda_local_model" >>> localModel.save(local_model_path) >>> sameLocalModel = LocalLDAModel.load(local_model_path)
New in version 2.0.0.
-
getDocConcentration()[source]¶ Gets the value of
docConcentrationor its default value.New in version 2.0.0.
-
getKeepLastCheckpoint()[source]¶ Gets the value of
keepLastCheckpointor its default value.New in version 2.0.0.
-
getLearningDecay()[source]¶ Gets the value of
learningDecayor its default value.New in version 2.0.0.
-
getLearningOffset()[source]¶ Gets the value of
learningOffsetor its default value.New in version 2.0.0.
-
getOptimizeDocConcentration()[source]¶ Gets the value of
optimizeDocConcentrationor its default value.New in version 2.0.0.
-
getSubsamplingRate()[source]¶ Gets the value of
subsamplingRateor its default value.New in version 2.0.0.
-
getTopicConcentration()[source]¶ Gets the value of
topicConcentrationor its default value.New in version 2.0.0.
-
getTopicDistributionCol()[source]¶ Gets the value of
topicDistributionColor its default value.New in version 2.0.0.
-
setDocConcentration(value)[source]¶ Sets the value of
docConcentration.>>> algo = LDA().setDocConcentration([0.1, 0.2]) >>> algo.getDocConcentration() [0.1..., 0.2...]
New in version 2.0.0.
-
setK(value)[source]¶ Sets the value of
k.>>> algo = LDA().setK(10) >>> algo.getK() 10
New in version 2.0.0.
-
setKeepLastCheckpoint(value)[source]¶ Sets the value of
keepLastCheckpoint.>>> algo = LDA().setKeepLastCheckpoint(False) >>> algo.getKeepLastCheckpoint() False
New in version 2.0.0.
-
setLearningDecay(value)[source]¶ Sets the value of
learningDecay.>>> algo = LDA().setLearningDecay(0.1) >>> algo.getLearningDecay() 0.1...
New in version 2.0.0.
-
setLearningOffset(value)[source]¶ Sets the value of
learningOffset.>>> algo = LDA().setLearningOffset(100) >>> algo.getLearningOffset() 100.0
New in version 2.0.0.
-
setOptimizeDocConcentration(value)[source]¶ Sets the value of
optimizeDocConcentration.>>> algo = LDA().setOptimizeDocConcentration(True) >>> algo.getOptimizeDocConcentration() True
New in version 2.0.0.
-
setOptimizer(value)[source]¶ Sets the value of
optimizer. Currently only support ‘em’ and ‘online’.>>> algo = LDA().setOptimizer("em") >>> algo.getOptimizer() 'em'
New in version 2.0.0.
-
setParams(self, featuresCol='features', maxIter=20, seed=None, checkpointInterval=10, k=10, optimizer='online', learningOffset=1024.0, learningDecay=0.51, subsamplingRate=0.05, optimizeDocConcentration=True, docConcentration=None, topicConcentration=None, topicDistributionCol='topicDistribution', keepLastCheckpoint=True)[source]¶ Sets params for LDA.
New in version 2.0.0.
-
setSubsamplingRate(value)[source]¶ Sets the value of
subsamplingRate.>>> algo = LDA().setSubsamplingRate(0.1) >>> algo.getSubsamplingRate() 0.1...
New in version 2.0.0.
-
class
pyspark.ml.clustering.LDAModel(java_model=None)[source]¶ Latent Dirichlet Allocation (LDA) model. This abstraction permits for different underlying representations, including local and distributed data structures.
New in version 2.0.0.
-
describeTopics(maxTermsPerTopic=10)[source]¶ Return the topics described by their top-weighted terms.
New in version 2.0.0.
-
estimatedDocConcentration()[source]¶ Value for
LDA.docConcentrationestimated from data. If Online LDA was used andLDA.optimizeDocConcentrationwas set to false, then this returns the fixed (given) value for theLDA.docConcentrationparameter.New in version 2.0.0.
-
isDistributed()[source]¶ Indicates whether this instance is of type DistributedLDAModel
New in version 2.0.0.
-
logLikelihood(dataset)[source]¶ Calculates a lower bound on the log likelihood of the entire corpus. See Equation (16) in the Online LDA paper (Hoffman et al., 2010).
WARNING: If this model is an instance of
DistributedLDAModel(produced whenoptimizeris set to “em”), this involves collecting a largetopicsMatrix()to the driver. This implementation may be changed in the future.New in version 2.0.0.
-
logPerplexity(dataset)[source]¶ Calculate an upper bound on perplexity. (Lower is better.) See Equation (16) in the Online LDA paper (Hoffman et al., 2010).
WARNING: If this model is an instance of
DistributedLDAModel(produced whenoptimizeris set to “em”), this involves collecting a largetopicsMatrix()to the driver. This implementation may be changed in the future.New in version 2.0.0.
-
topicsMatrix()[source]¶ Inferred topics, where each topic is represented by a distribution over terms. This is a matrix of size vocabSize x k, where each column is a topic. No guarantees are given about the ordering of the topics.
WARNING: If this model is actually a
DistributedLDAModelinstance produced by the Expectation-Maximization (“em”) , then this method could involve
collecting a large amount of data to the driver (on the order of vocabSize x k).
, then this method could involve
collecting a large amount of data to the driver (on the order of vocabSize x k).New in version 2.0.0.
-
-
class
pyspark.ml.clustering.LocalLDAModel(java_model=None)[source]¶ Local (non-distributed) model fitted by
LDA. This model stores the inferred topics only; it does not store info about the training dataset.New in version 2.0.0.
-
class
pyspark.ml.clustering.PowerIterationClustering(*args, **kwargs)[source]¶ Note
Experimental
Power Iteration Clustering (PIC), a scalable graph clustering algorithm developed by Lin and Cohen. From the abstract: PIC finds a very low-dimensional embedding of a dataset using truncated power iteration on a normalized pair-wise similarity matrix of the data.
This class is not yet an Estimator/Transformer, use
assignClusters()method to run the PowerIterationClustering algorithm.See also
>>> data = [(1, 0, 0.5), ... (2, 0, 0.5), (2, 1, 0.7), ... (3, 0, 0.5), (3, 1, 0.7), (3, 2, 0.9), ... (4, 0, 0.5), (4, 1, 0.7), (4, 2, 0.9), (4, 3, 1.1), ... (5, 0, 0.5), (5, 1, 0.7), (5, 2, 0.9), (5, 3, 1.1), (5, 4, 1.3)] >>> df = spark.createDataFrame(data).toDF("src", "dst", "weight") >>> pic = PowerIterationClustering(k=2, maxIter=40, weightCol="weight") >>> assignments = pic.assignClusters(df) >>> assignments.sort(assignments.id).show(truncate=False) +---+-------+ |id |cluster| +---+-------+ |0 |1 | |1 |1 | |2 |1 | |3 |1 | |4 |1 | |5 |0 | +---+-------+ ... >>> pic_path = temp_path + "/pic" >>> pic.save(pic_path) >>> pic2 = PowerIterationClustering.load(pic_path) >>> pic2.getK() 2 >>> pic2.getMaxIter() 40
New in version 2.4.0.
-
assignClusters(dataset)[source]¶ Run the PIC algorithm and returns a cluster assignment for each input vertex.
- Parameters
dataset – A dataset with columns src, dst, weight representing the affinity matrix, which is the matrix A in the PIC paper. Suppose the src column value is i, the dst column value is j, the weight column value is similarity s,,ij,, which must be nonnegative. This is a symmetric matrix and hence s,,ij,, = s,,ji,,. For any (i, j) with nonzero similarity, there should be either (i, j, s,,ij,,) or (j, i, s,,ji,,) in the input. Rows with i = j are ignored, because we assume s,,ij,, = 0.0.
- Returns
A dataset that contains columns of vertex id and the corresponding cluster for the id. The schema of it will be: - id: Long - cluster: Int
New in version 2.4.0.
New in version 2.4.0.
-
27.5. Recommendation API¶
-
class
pyspark.ml.recommendation.ALS(*args, **kwargs)[source]¶ Alternating Least Squares (ALS) matrix factorization.
ALS attempts to estimate the ratings matrix
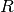 as the product of
two lower-rank matrices,
as the product of
two lower-rank matrices,  and
and  , i.e.
, i.e.  . Typically
these approximations are called ‘factor’ matrices. The general
approach is iterative. During each iteration, one of the factor
matrices is held constant, while the other is solved for using least
squares. The newly-solved factor matrix is then held constant while
solving for the other factor matrix.
. Typically
these approximations are called ‘factor’ matrices. The general
approach is iterative. During each iteration, one of the factor
matrices is held constant, while the other is solved for using least
squares. The newly-solved factor matrix is then held constant while
solving for the other factor matrix.This is a blocked implementation of the ALS factorization algorithm that groups the two sets of factors (referred to as “users” and “products”) into blocks and reduces communication by only sending one copy of each user vector to each product block on each iteration, and only for the product blocks that need that user’s feature vector. This is achieved by pre-computing some information about the ratings matrix to determine the “out-links” of each user (which blocks of products it will contribute to) and “in-link” information for each product (which of the feature vectors it receives from each user block it will depend on). This allows us to send only an array of feature vectors between each user block and product block, and have the product block find the users’ ratings and update the products based on these messages.
For implicit preference data, the algorithm used is based on “Collaborative Filtering for Implicit Feedback Datasets”,, adapted for the blocked approach used here.
Essentially instead of finding the low-rank approximations to the rating matrix
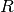 , this finds the approximations for a preference
matrix
, this finds the approximations for a preference
matrix 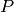 where the elements of
where the elements of 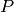 are 1 if r > 0 and 0 if r <= 0.
The ratings then act as ‘confidence’ values related to strength of
indicated user preferences rather than explicit ratings given to
items.
are 1 if r > 0 and 0 if r <= 0.
The ratings then act as ‘confidence’ values related to strength of
indicated user preferences rather than explicit ratings given to
items.>>> df = spark.createDataFrame( ... [(0, 0, 4.0), (0, 1, 2.0), (1, 1, 3.0), (1, 2, 4.0), (2, 1, 1.0), (2, 2, 5.0)], ... ["user", "item", "rating"]) >>> als = ALS(rank=10, maxIter=5, seed=0) >>> model = als.fit(df) >>> model.rank 10 >>> model.userFactors.orderBy("id").collect() [Row(id=0, features=[...]), Row(id=1, ...), Row(id=2, ...)] >>> test = spark.createDataFrame([(0, 2), (1, 0), (2, 0)], ["user", "item"]) >>> predictions = sorted(model.transform(test).collect(), key=lambda r: r[0]) >>> predictions[0] Row(user=0, item=2, prediction=-0.13807615637779236) >>> predictions[1] Row(user=1, item=0, prediction=2.6258413791656494) >>> predictions[2] Row(user=2, item=0, prediction=-1.5018409490585327) >>> user_recs = model.recommendForAllUsers(3) >>> user_recs.where(user_recs.user == 0) .select("recommendations.item", "recommendations.rating").collect() [Row(item=[0, 1, 2], rating=[3.910..., 1.992..., -0.138...])] >>> item_recs = model.recommendForAllItems(3) >>> item_recs.where(item_recs.item == 2) .select("recommendations.user", "recommendations.rating").collect() [Row(user=[2, 1, 0], rating=[4.901..., 3.981..., -0.138...])] >>> user_subset = df.where(df.user == 2) >>> user_subset_recs = model.recommendForUserSubset(user_subset, 3) >>> user_subset_recs.select("recommendations.item", "recommendations.rating").first() Row(item=[2, 1, 0], rating=[4.901..., 1.056..., -1.501...]) >>> item_subset = df.where(df.item == 0) >>> item_subset_recs = model.recommendForItemSubset(item_subset, 3) >>> item_subset_recs.select("recommendations.user", "recommendations.rating").first() Row(user=[0, 1, 2], rating=[3.910..., 2.625..., -1.501...]) >>> als_path = temp_path + "/als" >>> als.save(als_path) >>> als2 = ALS.load(als_path) >>> als.getMaxIter() 5 >>> model_path = temp_path + "/als_model" >>> model.save(model_path) >>> model2 = ALSModel.load(model_path) >>> model.rank == model2.rank True >>> sorted(model.userFactors.collect()) == sorted(model2.userFactors.collect()) True >>> sorted(model.itemFactors.collect()) == sorted(model2.itemFactors.collect()) True
New in version 1.4.0.
-
getColdStartStrategy()[source]¶ Gets the value of coldStartStrategy or its default value.
New in version 2.2.0.
-
getFinalStorageLevel()[source]¶ Gets the value of finalStorageLevel or its default value.
New in version 2.0.0.
-
getImplicitPrefs()[source]¶ Gets the value of implicitPrefs or its default value.
New in version 1.4.0.
-
getIntermediateStorageLevel()[source]¶ Gets the value of intermediateStorageLevel or its default value.
New in version 2.0.0.
-
getNumItemBlocks()[source]¶ Gets the value of numItemBlocks or its default value.
New in version 1.4.0.
-
getNumUserBlocks()[source]¶ Gets the value of numUserBlocks or its default value.
New in version 1.4.0.
-
setIntermediateStorageLevel(value)[source]¶ Sets the value of
intermediateStorageLevel.New in version 2.0.0.
-
setNumBlocks(value)[source]¶ Sets both
numUserBlocksandnumItemBlocksto the specific value.New in version 1.4.0.
-
setParams(self, rank=10, maxIter=10, regParam=0.1, numUserBlocks=10, numItemBlocks=10, implicitPrefs=False, alpha=1.0, userCol='user', itemCol='item', seed=None, ratingCol='rating', nonnegative=False, checkpointInterval=10, intermediateStorageLevel='MEMORY_AND_DISK', finalStorageLevel='MEMORY_AND_DISK', coldStartStrategy='nan')[source]¶ Sets params for ALS.
New in version 1.4.0.
-
-
class
pyspark.ml.recommendation.ALSModel(java_model=None)[source]¶ Model fitted by ALS.
New in version 1.4.0.
-
property
itemFactors[source]¶ 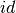 and
and
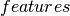
New in version 1.4.0.
- Type
a DataFrame that stores item factors in two columns
-
recommendForAllItems(numUsers)[source]¶ Returns top
 users recommended for each item, for all items.
users recommended for each item, for all items.- Parameters
numUsers – max number of recommendations for each item
- Returns
a DataFrame of (itemCol, recommendations), where recommendations are stored as an array of (userCol, rating) Rows.
New in version 2.2.0.
-
recommendForAllUsers(numItems)[source]¶ Returns top
 items recommended for each user, for all users.
items recommended for each user, for all users.- Parameters
numItems – max number of recommendations for each user
- Returns
a DataFrame of (userCol, recommendations), where recommendations are stored as an array of (itemCol, rating) Rows.
New in version 2.2.0.
-
recommendForItemSubset(dataset, numUsers)[source]¶ Returns top
 users recommended for each item id in the input data set. Note that
if there are duplicate ids in the input dataset, only one set of recommendations per unique
id will be returned.
users recommended for each item id in the input data set. Note that
if there are duplicate ids in the input dataset, only one set of recommendations per unique
id will be returned.- Parameters
dataset – a Dataset containing a column of item ids. The column name must match
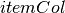 .
.numUsers – max number of recommendations for each item
- Returns
a DataFrame of (itemCol, recommendations), where recommendations are stored as an array of (userCol, rating) Rows.
New in version 2.3.0.
-
recommendForUserSubset(dataset, numItems)[source]¶ Returns top
 items recommended for each user id in the input data set. Note that
if there are duplicate ids in the input dataset, only one set of recommendations per unique
id will be returned.
items recommended for each user id in the input data set. Note that
if there are duplicate ids in the input dataset, only one set of recommendations per unique
id will be returned.- Parameters
dataset – a Dataset containing a column of user ids. The column name must match
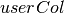 .
.numItems – max number of recommendations for each user
- Returns
a DataFrame of (userCol, recommendations), where recommendations are stored as an array of (itemCol, rating) Rows.
New in version 2.3.0.
-
property
27.6. Pipeline API¶
-
class
pyspark.ml.pipeline.Pipeline(*args, **kwargs)[source]¶ A simple pipeline, which acts as an estimator. A Pipeline consists of a sequence of stages, each of which is either an
Estimatoror aTransformer. WhenPipeline.fit()is called, the stages are executed in order. If a stage is anEstimator, itsEstimator.fit()method will be called on the input dataset to fit a model. Then the model, which is a transformer, will be used to transform the dataset as the input to the next stage. If a stage is aTransformer, itsTransformer.transform()method will be called to produce the dataset for the next stage. The fitted model from aPipelineis aPipelineModel, which consists of fitted models and transformers, corresponding to the pipeline stages. If stages is an empty list, the pipeline acts as an identity transformer.New in version 1.3.0.
-
copy(extra=None)[source]¶ Creates a copy of this instance.
- Parameters
extra – extra parameters
- Returns
new instance
New in version 1.4.0.
-
-
class
pyspark.ml.pipeline.PipelineModel(stages)[source]¶ Represents a compiled pipeline with transformers and fitted models.
New in version 1.3.0.
-
class
pyspark.ml.pipeline.PipelineModelReader(cls)[source]¶ (Private) Specialization of
MLReaderforPipelineModeltypes
-
class
pyspark.ml.pipeline.PipelineModelWriter(instance)[source]¶ (Private) Specialization of
MLWriterforPipelineModeltypes
-
class
pyspark.ml.pipeline.PipelineReader(cls)[source]¶ (Private) Specialization of
MLReaderforPipelinetypes
Note
DeveloperApi
Functions for
MLReaderandMLWritershared betweenPipelineandPipelineModelNew in version 2.3.0.
Get path for saving the given stage.
Load metadata and stages for a
PipelineorPipelineModel- Returns
(UID, list of stages)
Save metadata and stages for a
PipelineorPipelineModel- save metadata to path/metadata - save stages to stages/IDX_UID
Check that all stages are Writable
27.7. Tuning API¶
-
class
pyspark.ml.tuning.CrossValidator(*args, **kwargs)[source]¶ K-fold cross validation performs model selection by splitting the dataset into a set of non-overlapping randomly partitioned folds which are used as separate training and test datasets e.g., with k=3 folds, K-fold cross validation will generate 3 (training, test) dataset pairs, each of which uses 2/3 of the data for training and 1/3 for testing. Each fold is used as the test set exactly once.
>>> from pyspark.ml.classification import LogisticRegression >>> from pyspark.ml.evaluation import BinaryClassificationEvaluator >>> from pyspark.ml.linalg import Vectors >>> dataset = spark.createDataFrame( ... [(Vectors.dense([0.0]), 0.0), ... (Vectors.dense([0.4]), 1.0), ... (Vectors.dense([0.5]), 0.0), ... (Vectors.dense([0.6]), 1.0), ... (Vectors.dense([1.0]), 1.0)] * 10, ... ["features", "label"]) >>> lr = LogisticRegression() >>> grid = ParamGridBuilder().addGrid(lr.maxIter, [0, 1]).build() >>> evaluator = BinaryClassificationEvaluator() >>> cv = CrossValidator(estimator=lr, estimatorParamMaps=grid, evaluator=evaluator, ... parallelism=2) >>> cvModel = cv.fit(dataset) >>> cvModel.avgMetrics[0] 0.5 >>> evaluator.evaluate(cvModel.transform(dataset)) 0.8333...
New in version 1.4.0.
-
copy(extra=None)[source]¶ Creates a copy of this instance with a randomly generated uid and some extra params. This copies creates a deep copy of the embedded paramMap, and copies the embedded and extra parameters over.
- Parameters
extra – Extra parameters to copy to the new instance
- Returns
Copy of this instance
New in version 1.4.0.
-
setParams(estimator=None, estimatorParamMaps=None, evaluator=None, numFolds=3, seed=None, parallelism=1, collectSubModels=False)[source]¶ setParams(self, estimator=None, estimatorParamMaps=None, evaluator=None, numFolds=3, seed=None, parallelism=1, collectSubModels=False): Sets params for cross validator.
New in version 1.4.0.
-
-
class
pyspark.ml.tuning.CrossValidatorModel(bestModel, avgMetrics=[], subModels=None)[source]¶ CrossValidatorModel contains the model with the highest average cross-validation metric across folds and uses this model to transform input data. CrossValidatorModel also tracks the metrics for each param map evaluated.
New in version 1.4.0.
-
avgMetrics[source]¶ Average cross-validation metrics for each paramMap in CrossValidator.estimatorParamMaps, in the corresponding order.
-
copy(extra=None)[source]¶ Creates a copy of this instance with a randomly generated uid and some extra params. This copies the underlying bestModel, creates a deep copy of the embedded paramMap, and copies the embedded and extra parameters over. It does not copy the extra Params into the subModels.
- Parameters
extra – Extra parameters to copy to the new instance
- Returns
Copy of this instance
New in version 1.4.0.
-
-
class
pyspark.ml.tuning.ParamGridBuilder[source]¶ Builder for a param grid used in grid search-based model selection.
>>> from pyspark.ml.classification import LogisticRegression >>> lr = LogisticRegression() >>> output = ParamGridBuilder() \ ... .baseOn({lr.labelCol: 'l'}) \ ... .baseOn([lr.predictionCol, 'p']) \ ... .addGrid(lr.regParam, [1.0, 2.0]) \ ... .addGrid(lr.maxIter, [1, 5]) \ ... .build() >>> expected = [ ... {lr.regParam: 1.0, lr.maxIter: 1, lr.labelCol: 'l', lr.predictionCol: 'p'}, ... {lr.regParam: 2.0, lr.maxIter: 1, lr.labelCol: 'l', lr.predictionCol: 'p'}, ... {lr.regParam: 1.0, lr.maxIter: 5, lr.labelCol: 'l', lr.predictionCol: 'p'}, ... {lr.regParam: 2.0, lr.maxIter: 5, lr.labelCol: 'l', lr.predictionCol: 'p'}] >>> len(output) == len(expected) True >>> all([m in expected for m in output]) True
New in version 1.4.0.
-
addGrid(param, values)[source]¶ Sets the given parameters in this grid to fixed values.
New in version 1.4.0.
-
-
class
pyspark.ml.tuning.TrainValidationSplit(*args, **kwargs)[source]¶ Note
Experimental
Validation for hyper-parameter tuning. Randomly splits the input dataset into train and validation sets, and uses evaluation metric on the validation set to select the best model. Similar to
CrossValidator, but only splits the set once.>>> from pyspark.ml.classification import LogisticRegression >>> from pyspark.ml.evaluation import BinaryClassificationEvaluator >>> from pyspark.ml.linalg import Vectors >>> dataset = spark.createDataFrame( ... [(Vectors.dense([0.0]), 0.0), ... (Vectors.dense([0.4]), 1.0), ... (Vectors.dense([0.5]), 0.0), ... (Vectors.dense([0.6]), 1.0), ... (Vectors.dense([1.0]), 1.0)] * 10, ... ["features", "label"]) >>> lr = LogisticRegression() >>> grid = ParamGridBuilder().addGrid(lr.maxIter, [0, 1]).build() >>> evaluator = BinaryClassificationEvaluator() >>> tvs = TrainValidationSplit(estimator=lr, estimatorParamMaps=grid, evaluator=evaluator, ... parallelism=2) >>> tvsModel = tvs.fit(dataset) >>> evaluator.evaluate(tvsModel.transform(dataset)) 0.8333...
New in version 2.0.0.
-
copy(extra=None)[source]¶ Creates a copy of this instance with a randomly generated uid and some extra params. This copies creates a deep copy of the embedded paramMap, and copies the embedded and extra parameters over.
- Parameters
extra – Extra parameters to copy to the new instance
- Returns
Copy of this instance
New in version 2.0.0.
-
setParams(estimator=None, estimatorParamMaps=None, evaluator=None, trainRatio=0.75, parallelism=1, collectSubModels=False, seed=None)[source]¶ setParams(self, estimator=None, estimatorParamMaps=None, evaluator=None, trainRatio=0.75, parallelism=1, collectSubModels=False, seed=None): Sets params for the train validation split.
New in version 2.0.0.
-
-
class
pyspark.ml.tuning.TrainValidationSplitModel(bestModel, validationMetrics=[], subModels=None)[source]¶ Note
Experimental
Model from train validation split.
New in version 2.0.0.
-
copy(extra=None)[source]¶ Creates a copy of this instance with a randomly generated uid and some extra params. This copies the underlying bestModel, creates a deep copy of the embedded paramMap, and copies the embedded and extra parameters over. And, this creates a shallow copy of the validationMetrics. It does not copy the extra Params into the subModels.
- Parameters
extra – Extra parameters to copy to the new instance
- Returns
Copy of this instance
New in version 2.0.0.
-
27.8. Evaluation API¶
-
class
pyspark.ml.evaluation.BinaryClassificationEvaluator(*args, **kwargs)[source]¶ Note
Experimental
Evaluator for binary classification, which expects two input columns: rawPrediction and label. The rawPrediction column can be of type double (binary 0/1 prediction, or probability of label 1) or of type vector (length-2 vector of raw predictions, scores, or label probabilities).
>>> from pyspark.ml.linalg import Vectors >>> scoreAndLabels = map(lambda x: (Vectors.dense([1.0 - x[0], x[0]]), x[1]), ... [(0.1, 0.0), (0.1, 1.0), (0.4, 0.0), (0.6, 0.0), (0.6, 1.0), (0.6, 1.0), (0.8, 1.0)]) >>> dataset = spark.createDataFrame(scoreAndLabels, ["raw", "label"]) ... >>> evaluator = BinaryClassificationEvaluator(rawPredictionCol="raw") >>> evaluator.evaluate(dataset) 0.70... >>> evaluator.evaluate(dataset, {evaluator.metricName: "areaUnderPR"}) 0.83... >>> bce_path = temp_path + "/bce" >>> evaluator.save(bce_path) >>> evaluator2 = BinaryClassificationEvaluator.load(bce_path) >>> str(evaluator2.getRawPredictionCol()) 'raw'
New in version 1.4.0.
-
class
pyspark.ml.evaluation.ClusteringEvaluator(*args, **kwargs)[source]¶ Note
Experimental
Evaluator for Clustering results, which expects two input columns: prediction and features. The metric computes the Silhouette measure using the squared Euclidean distance.
The Silhouette is a measure for the validation of the consistency within clusters. It ranges between 1 and -1, where a value close to 1 means that the points in a cluster are close to the other points in the same cluster and far from the points of the other clusters.
>>> from pyspark.ml.linalg import Vectors >>> featureAndPredictions = map(lambda x: (Vectors.dense(x[0]), x[1]), ... [([0.0, 0.5], 0.0), ([0.5, 0.0], 0.0), ([10.0, 11.0], 1.0), ... ([10.5, 11.5], 1.0), ([1.0, 1.0], 0.0), ([8.0, 6.0], 1.0)]) >>> dataset = spark.createDataFrame(featureAndPredictions, ["features", "prediction"]) ... >>> evaluator = ClusteringEvaluator(predictionCol="prediction") >>> evaluator.evaluate(dataset) 0.9079... >>> ce_path = temp_path + "/ce" >>> evaluator.save(ce_path) >>> evaluator2 = ClusteringEvaluator.load(ce_path) >>> str(evaluator2.getPredictionCol()) 'prediction'
New in version 2.3.0.
-
class
pyspark.ml.evaluation.Evaluator[source]¶ Base class for evaluators that compute metrics from predictions.
New in version 1.4.0.
-
evaluate(dataset, params=None)[source]¶ Evaluates the output with optional parameters.
- Parameters
dataset – a dataset that contains labels/observations and predictions
params – an optional param map that overrides embedded params
- Returns
metric
New in version 1.4.0.
-
isLargerBetter()[source]¶ Indicates whether the metric returned by
evaluate()should be maximized (True, default) or minimized (False). A given evaluator may support multiple metrics which may be maximized or minimized.New in version 1.5.0.
-
-
class
pyspark.ml.evaluation.MulticlassClassificationEvaluator(*args, **kwargs)[source]¶ Note
Experimental
Evaluator for Multiclass Classification, which expects two input columns: prediction and label.
>>> scoreAndLabels = [(0.0, 0.0), (0.0, 1.0), (0.0, 0.0), ... (1.0, 0.0), (1.0, 1.0), (1.0, 1.0), (1.0, 1.0), (2.0, 2.0), (2.0, 0.0)] >>> dataset = spark.createDataFrame(scoreAndLabels, ["prediction", "label"]) ... >>> evaluator = MulticlassClassificationEvaluator(predictionCol="prediction") >>> evaluator.evaluate(dataset) 0.66... >>> evaluator.evaluate(dataset, {evaluator.metricName: "accuracy"}) 0.66... >>> mce_path = temp_path + "/mce" >>> evaluator.save(mce_path) >>> evaluator2 = MulticlassClassificationEvaluator.load(mce_path) >>> str(evaluator2.getPredictionCol()) 'prediction'
New in version 1.5.0.
-
class
pyspark.ml.evaluation.RegressionEvaluator(*args, **kwargs)[source]¶ Note
Experimental
Evaluator for Regression, which expects two input columns: prediction and label.
>>> scoreAndLabels = [(-28.98343821, -27.0), (20.21491975, 21.5), ... (-25.98418959, -22.0), (30.69731842, 33.0), (74.69283752, 71.0)] >>> dataset = spark.createDataFrame(scoreAndLabels, ["raw", "label"]) ... >>> evaluator = RegressionEvaluator(predictionCol="raw") >>> evaluator.evaluate(dataset) 2.842... >>> evaluator.evaluate(dataset, {evaluator.metricName: "r2"}) 0.993... >>> evaluator.evaluate(dataset, {evaluator.metricName: "mae"}) 2.649... >>> re_path = temp_path + "/re" >>> evaluator.save(re_path) >>> evaluator2 = RegressionEvaluator.load(re_path) >>> str(evaluator2.getPredictionCol()) 'raw'
New in version 1.4.0.
 .
.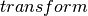 method.
method.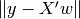 is scaled down when loss is “huber”, otherwise 1.0.
is scaled down when loss is “huber”, otherwise 1.0.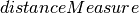 or its default value.
or its default value.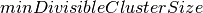 or its default value.
or its default value.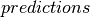 .
.