11. Classification¶
Chinese proverb
Birds of a feather folock together. – old Chinese proverb
11.1. Binomial logistic regression¶
11.1.1. Introduction¶
11.1.2. Demo¶
The Jupyter notebook can be download from Logistic Regression.
For more details, please visit Logistic Regression API .
Note
In this demo, I introduced a new function get_dummy to deal with the categorical data. I highly recommend you to use my get_dummy function in the other cases. This function will save a lot of time for you.
Set up spark context and SparkSession
from pyspark.sql import SparkSession
spark = SparkSession \
.builder \
.appName("Python Spark Logistic Regression example") \
.config("spark.some.config.option", "some-value") \
.getOrCreate()
Load dataset
df = spark.read.format('com.databricks.spark.csv') \
.options(header='true', inferschema='true') \
.load("./data/bank.csv",header=True);
df.drop('day','month','poutcome').show(5)
+---+------------+-------+---------+-------+-------+-------+----+-------+--------+--------+-----+--------+---+
|age| job|marital|education|default|balance|housing|loan|contact|duration|campaign|pdays|previous| y|
+---+------------+-------+---------+-------+-------+-------+----+-------+--------+--------+-----+--------+---+
| 58| management|married| tertiary| no| 2143| yes| no|unknown| 261| 1| -1| 0| no|
| 44| technician| single|secondary| no| 29| yes| no|unknown| 151| 1| -1| 0| no|
| 33|entrepreneur|married|secondary| no| 2| yes| yes|unknown| 76| 1| -1| 0| no|
| 47| blue-collar|married| unknown| no| 1506| yes| no|unknown| 92| 1| -1| 0| no|
| 33| unknown| single| unknown| no| 1| no| no|unknown| 198| 1| -1| 0| no|
+---+------------+-------+---------+-------+-------+-------+----+-------+--------+--------+-----+--------+---+
only showing top 5 rows
df.printSchema()
root
|-- age: integer (nullable = true)
|-- job: string (nullable = true)
|-- marital: string (nullable = true)
|-- education: string (nullable = true)
|-- default: string (nullable = true)
|-- balance: integer (nullable = true)
|-- housing: string (nullable = true)
|-- loan: string (nullable = true)
|-- contact: string (nullable = true)
|-- day: integer (nullable = true)
|-- month: string (nullable = true)
|-- duration: integer (nullable = true)
|-- campaign: integer (nullable = true)
|-- pdays: integer (nullable = true)
|-- previous: integer (nullable = true)
|-- poutcome: string (nullable = true)
|-- y: string (nullable = true)
Note
You are strongly encouraged to try my
get_dummyfunction for dealing with the categorical data in complex dataset.Supervised learning version:
def get_dummy(df,indexCol,categoricalCols,continuousCols,labelCol): from pyspark.ml import Pipeline from pyspark.ml.feature import StringIndexer, OneHotEncoder, VectorAssembler from pyspark.sql.functions import col indexers = [ StringIndexer(inputCol=c, outputCol="{0}_indexed".format(c)) for c in categoricalCols ] # default setting: dropLast=True encoders = [ OneHotEncoder(inputCol=indexer.getOutputCol(), outputCol="{0}_encoded".format(indexer.getOutputCol())) for indexer in indexers ] assembler = VectorAssembler(inputCols=[encoder.getOutputCol() for encoder in encoders] + continuousCols, outputCol="features") pipeline = Pipeline(stages=indexers + encoders + [assembler]) model=pipeline.fit(df) data = model.transform(df) data = data.withColumn('label',col(labelCol)) return data.select(indexCol,'features','label')Unsupervised learning version:
def get_dummy(df,indexCol,categoricalCols,continuousCols): ''' Get dummy variables and concat with continuous variables for unsupervised learning. :param df: the dataframe :param categoricalCols: the name list of the categorical data :param continuousCols: the name list of the numerical data :return k: feature matrix :author: Wenqiang Feng :email: von198@gmail.com ''' indexers = [ StringIndexer(inputCol=c, outputCol="{0}_indexed".format(c)) for c in categoricalCols ] # default setting: dropLast=True encoders = [ OneHotEncoder(inputCol=indexer.getOutputCol(), outputCol="{0}_encoded".format(indexer.getOutputCol())) for indexer in indexers ] assembler = VectorAssembler(inputCols=[encoder.getOutputCol() for encoder in encoders] + continuousCols, outputCol="features") pipeline = Pipeline(stages=indexers + encoders + [assembler]) model=pipeline.fit(df) data = model.transform(df) return data.select(indexCol,'features')
Two in one:
def get_dummy(df,indexCol,categoricalCols,continuousCols,labelCol,dropLast=False): ''' Get dummy variables and concat with continuous variables for ml modeling. :param df: the dataframe :param categoricalCols: the name list of the categorical data :param continuousCols: the name list of the numerical data :param labelCol: the name of label column :param dropLast: the flag of drop last column :return: feature matrix :author: Wenqiang Feng :email: von198@gmail.com >>> df = spark.createDataFrame([ (0, "a"), (1, "b"), (2, "c"), (3, "a"), (4, "a"), (5, "c") ], ["id", "category"]) >>> indexCol = 'id' >>> categoricalCols = ['category'] >>> continuousCols = [] >>> labelCol = [] >>> mat = get_dummy(df,indexCol,categoricalCols,continuousCols,labelCol) >>> mat.show() >>> +---+-------------+ | id| features| +---+-------------+ | 0|[1.0,0.0,0.0]| | 1|[0.0,0.0,1.0]| | 2|[0.0,1.0,0.0]| | 3|[1.0,0.0,0.0]| | 4|[1.0,0.0,0.0]| | 5|[0.0,1.0,0.0]| +---+-------------+ ''' from pyspark.ml import Pipeline from pyspark.ml.feature import StringIndexer, OneHotEncoder, VectorAssembler from pyspark.sql.functions import col indexers = [ StringIndexer(inputCol=c, outputCol="{0}_indexed".format(c)) for c in categoricalCols ] # default setting: dropLast=True encoders = [ OneHotEncoder(inputCol=indexer.getOutputCol(), outputCol="{0}_encoded".format(indexer.getOutputCol()),dropLast=dropLast) for indexer in indexers ] assembler = VectorAssembler(inputCols=[encoder.getOutputCol() for encoder in encoders] + continuousCols, outputCol="features") pipeline = Pipeline(stages=indexers + encoders + [assembler]) model=pipeline.fit(df) data = model.transform(df) if indexCol and labelCol: # for supervised learning data = data.withColumn('label',col(labelCol)) return data.select(indexCol,'features','label') elif not indexCol and labelCol: # for supervised learning data = data.withColumn('label',col(labelCol)) return data.select('features','label') elif indexCol and not labelCol: # for unsupervised learning return data.select(indexCol,'features') elif not indexCol and not labelCol: # for unsupervised learning return data.select('features')
def get_dummy(df,categoricalCols,continuousCols,labelCol):
from pyspark.ml import Pipeline
from pyspark.ml.feature import StringIndexer, OneHotEncoder, VectorAssembler
from pyspark.sql.functions import col
indexers = [ StringIndexer(inputCol=c, outputCol="{0}_indexed".format(c))
for c in categoricalCols ]
# default setting: dropLast=True
encoders = [ OneHotEncoder(inputCol=indexer.getOutputCol(),
outputCol="{0}_encoded".format(indexer.getOutputCol()))
for indexer in indexers ]
assembler = VectorAssembler(inputCols=[encoder.getOutputCol() for encoder in encoders]
+ continuousCols, outputCol="features")
pipeline = Pipeline(stages=indexers + encoders + [assembler])
model=pipeline.fit(df)
data = model.transform(df)
data = data.withColumn('label',col(labelCol))
return data.select('features','label')
Deal with categorical data and Convert the data to dense vector
catcols = ['job','marital','education','default',
'housing','loan','contact','poutcome']
num_cols = ['balance', 'duration','campaign','pdays','previous',]
labelCol = 'y'
data = get_dummy(df,catcols,num_cols,labelCol)
data.show(5)
+--------------------+-----+
| features|label|
+--------------------+-----+
|(29,[1,11,14,16,1...| no|
|(29,[2,12,13,16,1...| no|
|(29,[7,11,13,16,1...| no|
|(29,[0,11,16,17,1...| no|
|(29,[12,16,18,20,...| no|
+--------------------+-----+
only showing top 5 rows
Deal with Categorical Label and Variables
from pyspark.ml.feature import StringIndexer
# Index labels, adding metadata to the label column
labelIndexer = StringIndexer(inputCol='label',
outputCol='indexedLabel').fit(data)
labelIndexer.transform(data).show(5, True)
+--------------------+-----+------------+
| features|label|indexedLabel|
+--------------------+-----+------------+
|(29,[1,11,14,16,1...| no| 0.0|
|(29,[2,12,13,16,1...| no| 0.0|
|(29,[7,11,13,16,1...| no| 0.0|
|(29,[0,11,16,17,1...| no| 0.0|
|(29,[12,16,18,20,...| no| 0.0|
+--------------------+-----+------------+
only showing top 5 rows
from pyspark.ml.feature import VectorIndexer
# Automatically identify categorical features, and index them.
# Set maxCategories so features with > 4 distinct values are treated as continuous.
featureIndexer =VectorIndexer(inputCol="features", \
outputCol="indexedFeatures", \
maxCategories=4).fit(data)
featureIndexer.transform(data).show(5, True)
+--------------------+-----+--------------------+
| features|label| indexedFeatures|
+--------------------+-----+--------------------+
|(29,[1,11,14,16,1...| no|(29,[1,11,14,16,1...|
|(29,[2,12,13,16,1...| no|(29,[2,12,13,16,1...|
|(29,[7,11,13,16,1...| no|(29,[7,11,13,16,1...|
|(29,[0,11,16,17,1...| no|(29,[0,11,16,17,1...|
|(29,[12,16,18,20,...| no|(29,[12,16,18,20,...|
+--------------------+-----+--------------------+
only showing top 5 rows
Split the data to training and test data sets
# Split the data into training and test sets (40% held out for testing)
(trainingData, testData) = data.randomSplit([0.6, 0.4])
trainingData.show(5,False)
testData.show(5,False)
+-------------------------------------------------------------------------------------------------+-----+
|features |label|
+-------------------------------------------------------------------------------------------------+-----+
|(29,[0,11,13,16,17,18,19,21,24,25,26,27],[1.0,1.0,1.0,1.0,1.0,1.0,1.0,1.0,-731.0,401.0,4.0,-1.0])|no |
|(29,[0,11,13,16,17,18,19,21,24,25,26,27],[1.0,1.0,1.0,1.0,1.0,1.0,1.0,1.0,-723.0,112.0,2.0,-1.0])|no |
|(29,[0,11,13,16,17,18,19,21,24,25,26,27],[1.0,1.0,1.0,1.0,1.0,1.0,1.0,1.0,-626.0,205.0,1.0,-1.0])|no |
|(29,[0,11,13,16,17,18,19,21,24,25,26,27],[1.0,1.0,1.0,1.0,1.0,1.0,1.0,1.0,-498.0,357.0,1.0,-1.0])|no |
|(29,[0,11,13,16,17,18,19,21,24,25,26,27],[1.0,1.0,1.0,1.0,1.0,1.0,1.0,1.0,-477.0,473.0,2.0,-1.0])|no |
+-------------------------------------------------------------------------------------------------+-----+
only showing top 5 rows
+-------------------------------------------------------------------------------------------------+-----+
|features |label|
+-------------------------------------------------------------------------------------------------+-----+
|(29,[0,11,13,16,17,18,19,21,24,25,26,27],[1.0,1.0,1.0,1.0,1.0,1.0,1.0,1.0,-648.0,280.0,2.0,-1.0])|no |
|(29,[0,11,13,16,17,18,19,21,24,25,26,27],[1.0,1.0,1.0,1.0,1.0,1.0,1.0,1.0,-596.0,147.0,1.0,-1.0])|no |
|(29,[0,11,13,16,17,18,19,21,24,25,26,27],[1.0,1.0,1.0,1.0,1.0,1.0,1.0,1.0,-529.0,416.0,4.0,-1.0])|no |
|(29,[0,11,13,16,17,18,19,21,24,25,26,27],[1.0,1.0,1.0,1.0,1.0,1.0,1.0,1.0,-518.0,46.0,5.0,-1.0]) |no |
|(29,[0,11,13,16,17,18,19,21,24,25,26,27],[1.0,1.0,1.0,1.0,1.0,1.0,1.0,1.0,-470.0,275.0,2.0,-1.0])|no |
+-------------------------------------------------------------------------------------------------+-----+
only showing top 5 rows
Fit Logistic Regression Model
from pyspark.ml.classification import LogisticRegression
logr = LogisticRegression(featuresCol='indexedFeatures', labelCol='indexedLabel')
Pipeline Architecture
# Convert indexed labels back to original labels.
labelConverter = IndexToString(inputCol="prediction", outputCol="predictedLabel",
labels=labelIndexer.labels)
# Chain indexers and tree in a Pipeline
pipeline = Pipeline(stages=[labelIndexer, featureIndexer, logr,labelConverter])
# Train model. This also runs the indexers.
model = pipeline.fit(trainingData)
Make predictions
# Make predictions.
predictions = model.transform(testData)
# Select example rows to display.
predictions.select("features","label","predictedLabel").show(5)
+--------------------+-----+--------------+
| features|label|predictedLabel|
+--------------------+-----+--------------+
|(29,[0,11,13,16,1...| no| no|
|(29,[0,11,13,16,1...| no| no|
|(29,[0,11,13,16,1...| no| no|
|(29,[0,11,13,16,1...| no| no|
|(29,[0,11,13,16,1...| no| no|
+--------------------+-----+--------------+
only showing top 5 rows
Evaluation
from pyspark.ml.evaluation import MulticlassClassificationEvaluator
# Select (prediction, true label) and compute test error
evaluator = MulticlassClassificationEvaluator(
labelCol="indexedLabel", predictionCol="prediction", metricName="accuracy")
accuracy = evaluator.evaluate(predictions)
print("Test Error = %g" % (1.0 - accuracy))
Test Error = 0.0987688
lrModel = model.stages[2]
trainingSummary = lrModel.summary
# Obtain the objective per iteration
# objectiveHistory = trainingSummary.objectiveHistory
# print("objectiveHistory:")
# for objective in objectiveHistory:
# print(objective)
# Obtain the receiver-operating characteristic as a dataframe and areaUnderROC.
trainingSummary.roc.show(5)
print("areaUnderROC: " + str(trainingSummary.areaUnderROC))
# Set the model threshold to maximize F-Measure
fMeasure = trainingSummary.fMeasureByThreshold
maxFMeasure = fMeasure.groupBy().max('F-Measure').select('max(F-Measure)').head(5)
# bestThreshold = fMeasure.where(fMeasure['F-Measure'] == maxFMeasure['max(F-Measure)']) \
# .select('threshold').head()['threshold']
# lr.setThreshold(bestThreshold)
You can use z.show() to get the data and plot the ROC curves:
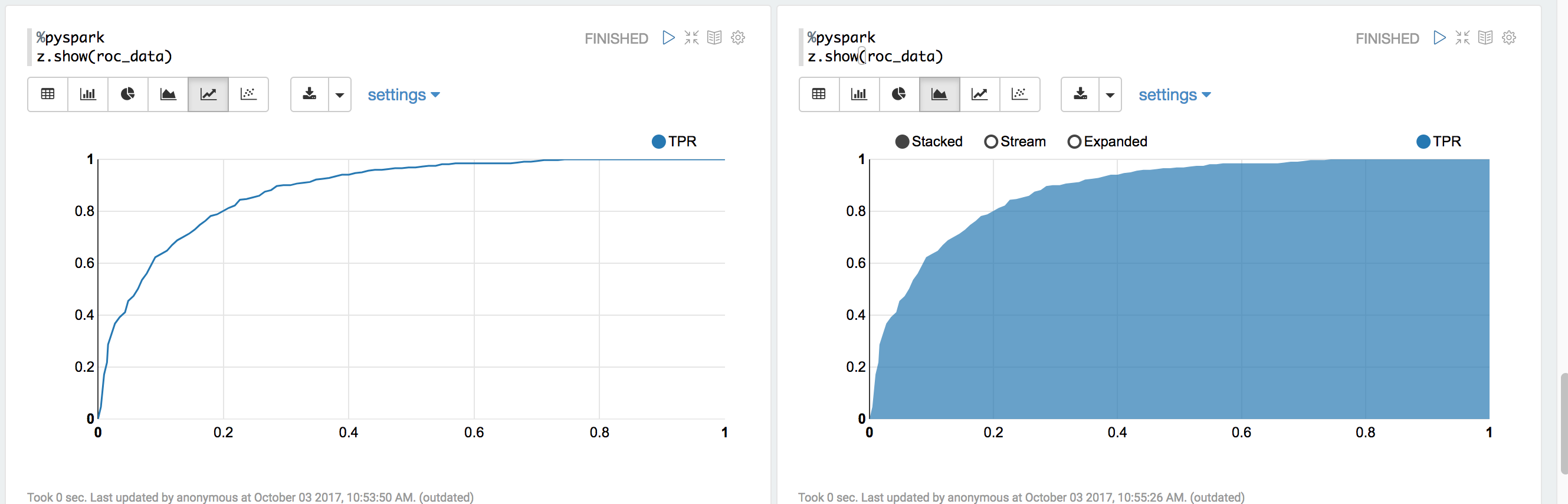
You can also register a TempTable data.registerTempTable('roc_data') and then
use sql to plot the ROC curve:
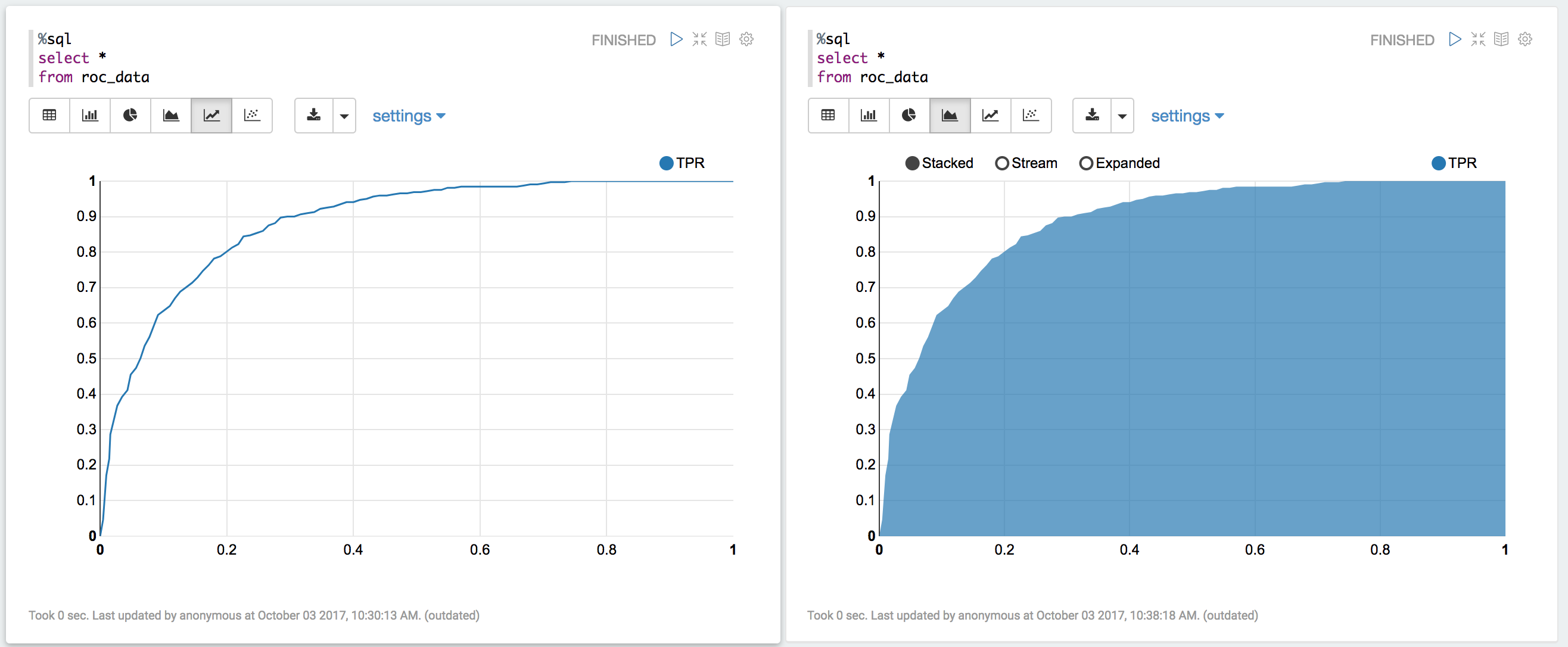
visualization
import matplotlib.pyplot as plt
import numpy as np
import itertools
def plot_confusion_matrix(cm, classes,
normalize=False,
title='Confusion matrix',
cmap=plt.cm.Blues):
"""
This function prints and plots the confusion matrix.
Normalization can be applied by setting `normalize=True`.
"""
if normalize:
cm = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]
print("Normalized confusion matrix")
else:
print('Confusion matrix, without normalization')
print(cm)
plt.imshow(cm, interpolation='nearest', cmap=cmap)
plt.title(title)
plt.colorbar()
tick_marks = np.arange(len(classes))
plt.xticks(tick_marks, classes, rotation=45)
plt.yticks(tick_marks, classes)
fmt = '.2f' if normalize else 'd'
thresh = cm.max() / 2.
for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):
plt.text(j, i, format(cm[i, j], fmt),
horizontalalignment="center",
color="white" if cm[i, j] > thresh else "black")
plt.tight_layout()
plt.ylabel('True label')
plt.xlabel('Predicted label')
class_temp = predictions.select("label").groupBy("label")\
.count().sort('count', ascending=False).toPandas()
class_temp = class_temp["label"].values.tolist()
class_names = map(str, class_temp)
# # # print(class_name)
class_names
['no', 'yes']
from sklearn.metrics import confusion_matrix
y_true = predictions.select("label")
y_true = y_true.toPandas()
y_pred = predictions.select("predictedLabel")
y_pred = y_pred.toPandas()
cnf_matrix = confusion_matrix(y_true, y_pred,labels=class_names)
cnf_matrix
array([[15657, 379],
[ 1410, 667]])
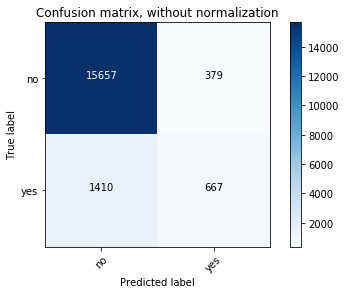
# Plot non-normalized confusion matrix
plt.figure()
plot_confusion_matrix(cnf_matrix, classes=class_names,
title='Confusion matrix, without normalization')
plt.show()
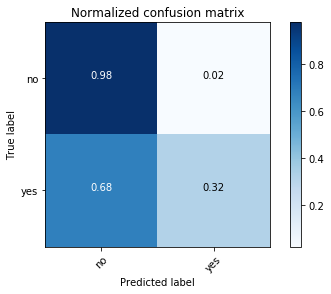
Confusion matrix, without normalization
[[15657 379]
[ 1410 667]]
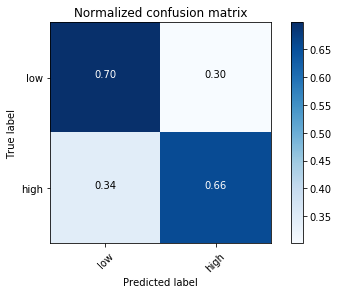
# Plot normalized confusion matrix
plt.figure()
plot_confusion_matrix(cnf_matrix, classes=class_names, normalize=True,
title='Normalized confusion matrix')
plt.show()
Normalized confusion matrix
[[ 0.97636568 0.02363432]
[ 0.67886375 0.32113625]]
11.2. Multinomial logistic regression¶
11.2.1. Introduction¶
11.2.2. Demo¶
The Jupyter notebook can be download from Logistic Regression.
For more details, please visit Logistic Regression API .
Note
In this demo, I introduced a new function get_dummy to deal with the categorical data. I highly recommend you to use my get_dummy function in the other cases. This function will save a lot of time for you.
Set up spark context and SparkSession
from pyspark.sql import SparkSession
spark = SparkSession \
.builder \
.appName("Python Spark MultinomialLogisticRegression classification") \
.config("spark.some.config.option", "some-value") \
.getOrCreate()
Load dataset
df = spark.read.format('com.databricks.spark.csv') \
.options(header='true', inferschema='true') \
.load("./data/WineData2.csv",header=True);
df.show(5)
+-----+--------+------+-----+---------+----+-----+-------+----+---------+-------+-------+
|fixed|volatile|citric|sugar|chlorides|free|total|density| pH|sulphates|alcohol|quality|
+-----+--------+------+-----+---------+----+-----+-------+----+---------+-------+-------+
| 7.4| 0.7| 0.0| 1.9| 0.076|11.0| 34.0| 0.9978|3.51| 0.56| 9.4| 5|
| 7.8| 0.88| 0.0| 2.6| 0.098|25.0| 67.0| 0.9968| 3.2| 0.68| 9.8| 5|
| 7.8| 0.76| 0.04| 2.3| 0.092|15.0| 54.0| 0.997|3.26| 0.65| 9.8| 5|
| 11.2| 0.28| 0.56| 1.9| 0.075|17.0| 60.0| 0.998|3.16| 0.58| 9.8| 6|
| 7.4| 0.7| 0.0| 1.9| 0.076|11.0| 34.0| 0.9978|3.51| 0.56| 9.4| 5|
+-----+--------+------+-----+---------+----+-----+-------+----+---------+-------+-------+
only showing top 5 rows
df.printSchema()
root
|-- fixed: double (nullable = true)
|-- volatile: double (nullable = true)
|-- citric: double (nullable = true)
|-- sugar: double (nullable = true)
|-- chlorides: double (nullable = true)
|-- free: double (nullable = true)
|-- total: double (nullable = true)
|-- density: double (nullable = true)
|-- pH: double (nullable = true)
|-- sulphates: double (nullable = true)
|-- alcohol: double (nullable = true)
|-- quality: string (nullable = true)
# Convert to float format
def string_to_float(x):
return float(x)
#
def condition(r):
if (0<= r <= 4):
label = "low"
elif(4< r <= 6):
label = "medium"
else:
label = "high"
return label
from pyspark.sql.functions import udf
from pyspark.sql.types import StringType, DoubleType
string_to_float_udf = udf(string_to_float, DoubleType())
quality_udf = udf(lambda x: condition(x), StringType())
df = df.withColumn("quality", quality_udf("quality"))
df.show(5,True)
+-----+--------+------+-----+---------+----+-----+-------+----+---------+-------+-------+
|fixed|volatile|citric|sugar|chlorides|free|total|density| pH|sulphates|alcohol|quality|
+-----+--------+------+-----+---------+----+-----+-------+----+---------+-------+-------+
| 7.4| 0.7| 0.0| 1.9| 0.076|11.0| 34.0| 0.9978|3.51| 0.56| 9.4| medium|
| 7.8| 0.88| 0.0| 2.6| 0.098|25.0| 67.0| 0.9968| 3.2| 0.68| 9.8| medium|
| 7.8| 0.76| 0.04| 2.3| 0.092|15.0| 54.0| 0.997|3.26| 0.65| 9.8| medium|
| 11.2| 0.28| 0.56| 1.9| 0.075|17.0| 60.0| 0.998|3.16| 0.58| 9.8| medium|
| 7.4| 0.7| 0.0| 1.9| 0.076|11.0| 34.0| 0.9978|3.51| 0.56| 9.4| medium|
+-----+--------+------+-----+---------+----+-----+-------+----+---------+-------+-------+
only showing top 5 rows
df.printSchema()
root
|-- fixed: double (nullable = true)
|-- volatile: double (nullable = true)
|-- citric: double (nullable = true)
|-- sugar: double (nullable = true)
|-- chlorides: double (nullable = true)
|-- free: double (nullable = true)
|-- total: double (nullable = true)
|-- density: double (nullable = true)
|-- pH: double (nullable = true)
|-- sulphates: double (nullable = true)
|-- alcohol: double (nullable = true)
|-- quality: string (nullable = true)
Deal with categorical data and Convert the data to dense vector
Note
You are strongly encouraged to try my
get_dummyfunction for dealing with the categorical data in complex dataset.Supervised learning version:
def get_dummy(df,indexCol,categoricalCols,continuousCols,labelCol): from pyspark.ml import Pipeline from pyspark.ml.feature import StringIndexer, OneHotEncoder, VectorAssembler from pyspark.sql.functions import col indexers = [ StringIndexer(inputCol=c, outputCol="{0}_indexed".format(c)) for c in categoricalCols ] # default setting: dropLast=True encoders = [ OneHotEncoder(inputCol=indexer.getOutputCol(), outputCol="{0}_encoded".format(indexer.getOutputCol())) for indexer in indexers ] assembler = VectorAssembler(inputCols=[encoder.getOutputCol() for encoder in encoders] + continuousCols, outputCol="features") pipeline = Pipeline(stages=indexers + encoders + [assembler]) model=pipeline.fit(df) data = model.transform(df) data = data.withColumn('label',col(labelCol)) return data.select(indexCol,'features','label')Unsupervised learning version:
def get_dummy(df,indexCol,categoricalCols,continuousCols): ''' Get dummy variables and concat with continuous variables for unsupervised learning. :param df: the dataframe :param categoricalCols: the name list of the categorical data :param continuousCols: the name list of the numerical data :return k: feature matrix :author: Wenqiang Feng :email: von198@gmail.com ''' indexers = [ StringIndexer(inputCol=c, outputCol="{0}_indexed".format(c)) for c in categoricalCols ] # default setting: dropLast=True encoders = [ OneHotEncoder(inputCol=indexer.getOutputCol(), outputCol="{0}_encoded".format(indexer.getOutputCol())) for indexer in indexers ] assembler = VectorAssembler(inputCols=[encoder.getOutputCol() for encoder in encoders] + continuousCols, outputCol="features") pipeline = Pipeline(stages=indexers + encoders + [assembler]) model=pipeline.fit(df) data = model.transform(df) return data.select(indexCol,'features')
Two in one:
def get_dummy(df,indexCol,categoricalCols,continuousCols,labelCol,dropLast=False): ''' Get dummy variables and concat with continuous variables for ml modeling. :param df: the dataframe :param categoricalCols: the name list of the categorical data :param continuousCols: the name list of the numerical data :param labelCol: the name of label column :param dropLast: the flag of drop last column :return: feature matrix :author: Wenqiang Feng :email: von198@gmail.com >>> df = spark.createDataFrame([ (0, "a"), (1, "b"), (2, "c"), (3, "a"), (4, "a"), (5, "c") ], ["id", "category"]) >>> indexCol = 'id' >>> categoricalCols = ['category'] >>> continuousCols = [] >>> labelCol = [] >>> mat = get_dummy(df,indexCol,categoricalCols,continuousCols,labelCol) >>> mat.show() >>> +---+-------------+ | id| features| +---+-------------+ | 0|[1.0,0.0,0.0]| | 1|[0.0,0.0,1.0]| | 2|[0.0,1.0,0.0]| | 3|[1.0,0.0,0.0]| | 4|[1.0,0.0,0.0]| | 5|[0.0,1.0,0.0]| +---+-------------+ ''' from pyspark.ml import Pipeline from pyspark.ml.feature import StringIndexer, OneHotEncoder, VectorAssembler from pyspark.sql.functions import col indexers = [ StringIndexer(inputCol=c, outputCol="{0}_indexed".format(c)) for c in categoricalCols ] # default setting: dropLast=True encoders = [ OneHotEncoder(inputCol=indexer.getOutputCol(), outputCol="{0}_encoded".format(indexer.getOutputCol()),dropLast=dropLast) for indexer in indexers ] assembler = VectorAssembler(inputCols=[encoder.getOutputCol() for encoder in encoders] + continuousCols, outputCol="features") pipeline = Pipeline(stages=indexers + encoders + [assembler]) model=pipeline.fit(df) data = model.transform(df) if indexCol and labelCol: # for supervised learning data = data.withColumn('label',col(labelCol)) return data.select(indexCol,'features','label') elif not indexCol and labelCol: # for supervised learning data = data.withColumn('label',col(labelCol)) return data.select('features','label') elif indexCol and not labelCol: # for unsupervised learning return data.select(indexCol,'features') elif not indexCol and not labelCol: # for unsupervised learning return data.select('features')
def get_dummy(df,categoricalCols,continuousCols,labelCol):
from pyspark.ml import Pipeline
from pyspark.ml.feature import StringIndexer, OneHotEncoder, VectorAssembler
from pyspark.sql.functions import col
indexers = [ StringIndexer(inputCol=c, outputCol="{0}_indexed".format(c))
for c in categoricalCols ]
# default setting: dropLast=True
encoders = [ OneHotEncoder(inputCol=indexer.getOutputCol(),
outputCol="{0}_encoded".format(indexer.getOutputCol()))
for indexer in indexers ]
assembler = VectorAssembler(inputCols=[encoder.getOutputCol() for encoder in encoders]
+ continuousCols, outputCol="features")
pipeline = Pipeline(stages=indexers + encoders + [assembler])
model=pipeline.fit(df)
data = model.transform(df)
data = data.withColumn('label',col(labelCol))
return data.select('features','label')
Transform the dataset to DataFrame
from pyspark.ml.linalg import Vectors # !!!!caution: not from pyspark.mllib.linalg import Vectors
from pyspark.ml import Pipeline
from pyspark.ml.feature import IndexToString,StringIndexer, VectorIndexer
from pyspark.ml.tuning import CrossValidator, ParamGridBuilder
from pyspark.ml.evaluation import MulticlassClassificationEvaluator
def transData(data):
return data.rdd.map(lambda r: [Vectors.dense(r[:-1]),r[-1]]).toDF(['features','label'])
transformed = transData(df)
transformed.show(5)
+--------------------+------+
| features| label|
+--------------------+------+
|[7.4,0.7,0.0,1.9,...|medium|
|[7.8,0.88,0.0,2.6...|medium|
|[7.8,0.76,0.04,2....|medium|
|[11.2,0.28,0.56,1...|medium|
|[7.4,0.7,0.0,1.9,...|medium|
+--------------------+------+
only showing top 5 rows
Deal with Categorical Label and Variables
# Index labels, adding metadata to the label column
labelIndexer = StringIndexer(inputCol='label',
outputCol='indexedLabel').fit(transformed)
labelIndexer.transform(transformed).show(5, True)
+--------------------+------+------------+
| features| label|indexedLabel|
+--------------------+------+------------+
|[7.4,0.7,0.0,1.9,...|medium| 0.0|
|[7.8,0.88,0.0,2.6...|medium| 0.0|
|[7.8,0.76,0.04,2....|medium| 0.0|
|[11.2,0.28,0.56,1...|medium| 0.0|
|[7.4,0.7,0.0,1.9,...|medium| 0.0|
+--------------------+------+------------+
only showing top 5 rows
# Automatically identify categorical features, and index them.
# Set maxCategories so features with > 4 distinct values are treated as continuous.
featureIndexer =VectorIndexer(inputCol="features", \
outputCol="indexedFeatures", \
maxCategories=4).fit(transformed)
featureIndexer.transform(transformed).show(5, True)
+--------------------+------+--------------------+
| features| label| indexedFeatures|
+--------------------+------+--------------------+
|[7.4,0.7,0.0,1.9,...|medium|[7.4,0.7,0.0,1.9,...|
|[7.8,0.88,0.0,2.6...|medium|[7.8,0.88,0.0,2.6...|
|[7.8,0.76,0.04,2....|medium|[7.8,0.76,0.04,2....|
|[11.2,0.28,0.56,1...|medium|[11.2,0.28,0.56,1...|
|[7.4,0.7,0.0,1.9,...|medium|[7.4,0.7,0.0,1.9,...|
+--------------------+------+--------------------+
only showing top 5 rows
Split the data to training and test data sets
# Split the data into training and test sets (40% held out for testing)
(trainingData, testData) = data.randomSplit([0.6, 0.4])
trainingData.show(5,False)
testData.show(5,False)
+---------------------------------------------------------+------+
|features |label |
+---------------------------------------------------------+------+
|[4.7,0.6,0.17,2.3,0.058,17.0,106.0,0.9932,3.85,0.6,12.9] |medium|
|[5.0,0.38,0.01,1.6,0.048,26.0,60.0,0.99084,3.7,0.75,14.0]|medium|
|[5.0,0.4,0.5,4.3,0.046,29.0,80.0,0.9902,3.49,0.66,13.6] |medium|
|[5.0,0.74,0.0,1.2,0.041,16.0,46.0,0.99258,4.01,0.59,12.5]|medium|
|[5.1,0.42,0.0,1.8,0.044,18.0,88.0,0.99157,3.68,0.73,13.6]|high |
+---------------------------------------------------------+------+
only showing top 5 rows
+---------------------------------------------------------+------+
|features |label |
+---------------------------------------------------------+------+
|[4.6,0.52,0.15,2.1,0.054,8.0,65.0,0.9934,3.9,0.56,13.1] |low |
|[4.9,0.42,0.0,2.1,0.048,16.0,42.0,0.99154,3.71,0.74,14.0]|high |
|[5.0,0.42,0.24,2.0,0.06,19.0,50.0,0.9917,3.72,0.74,14.0] |high |
|[5.0,1.02,0.04,1.4,0.045,41.0,85.0,0.9938,3.75,0.48,10.5]|low |
|[5.0,1.04,0.24,1.6,0.05,32.0,96.0,0.9934,3.74,0.62,11.5] |medium|
+---------------------------------------------------------+------+
only showing top 5 rows
Fit Multinomial logisticRegression Classification Model
from pyspark.ml.classification import LogisticRegression
logr = LogisticRegression(featuresCol='indexedFeatures', labelCol='indexedLabel')
Pipeline Architecture
# Convert indexed labels back to original labels.
labelConverter = IndexToString(inputCol="prediction", outputCol="predictedLabel",
labels=labelIndexer.labels)
# Chain indexers and tree in a Pipeline
pipeline = Pipeline(stages=[labelIndexer, featureIndexer, logr,labelConverter])
# Train model. This also runs the indexers.
model = pipeline.fit(trainingData)
Make predictions
# Make predictions.
predictions = model.transform(testData)
# Select example rows to display.
predictions.select("features","label","predictedLabel").show(5)
+--------------------+------+--------------+
| features| label|predictedLabel|
+--------------------+------+--------------+
|[4.6,0.52,0.15,2....| low| medium|
|[4.9,0.42,0.0,2.1...| high| high|
|[5.0,0.42,0.24,2....| high| high|
|[5.0,1.02,0.04,1....| low| medium|
|[5.0,1.04,0.24,1....|medium| medium|
+--------------------+------+--------------+
only showing top 5 rows
Evaluation
from pyspark.ml.evaluation import MulticlassClassificationEvaluator
# Select (prediction, true label) and compute test error
evaluator = MulticlassClassificationEvaluator(
labelCol="indexedLabel", predictionCol="prediction", metricName="accuracy")
accuracy = evaluator.evaluate(predictions)
print("Test Error = %g" % (1.0 - accuracy))
Test Error = 0.181287
lrModel = model.stages[2]
trainingSummary = lrModel.summary
# Obtain the objective per iteration
# objectiveHistory = trainingSummary.objectiveHistory
# print("objectiveHistory:")
# for objective in objectiveHistory:
# print(objective)
# Obtain the receiver-operating characteristic as a dataframe and areaUnderROC.
trainingSummary.roc.show(5)
print("areaUnderROC: " + str(trainingSummary.areaUnderROC))
# Set the model threshold to maximize F-Measure
fMeasure = trainingSummary.fMeasureByThreshold
maxFMeasure = fMeasure.groupBy().max('F-Measure').select('max(F-Measure)').head(5)
# bestThreshold = fMeasure.where(fMeasure['F-Measure'] == maxFMeasure['max(F-Measure)']) \
# .select('threshold').head()['threshold']
# lr.setThreshold(bestThreshold)
You can use z.show() to get the data and plot the ROC curves:

You can also register a TempTable data.registerTempTable('roc_data') and then
use sql to plot the ROC curve:

visualization
import matplotlib.pyplot as plt
import numpy as np
import itertools
def plot_confusion_matrix(cm, classes,
normalize=False,
title='Confusion matrix',
cmap=plt.cm.Blues):
"""
This function prints and plots the confusion matrix.
Normalization can be applied by setting `normalize=True`.
"""
if normalize:
cm = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]
print("Normalized confusion matrix")
else:
print('Confusion matrix, without normalization')
print(cm)
plt.imshow(cm, interpolation='nearest', cmap=cmap)
plt.title(title)
plt.colorbar()
tick_marks = np.arange(len(classes))
plt.xticks(tick_marks, classes, rotation=45)
plt.yticks(tick_marks, classes)
fmt = '.2f' if normalize else 'd'
thresh = cm.max() / 2.
for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):
plt.text(j, i, format(cm[i, j], fmt),
horizontalalignment="center",
color="white" if cm[i, j] > thresh else "black")
plt.tight_layout()
plt.ylabel('True label')
plt.xlabel('Predicted label')
class_temp = predictions.select("label").groupBy("label")\
.count().sort('count', ascending=False).toPandas()
class_temp = class_temp["label"].values.tolist()
class_names = map(str, class_temp)
# # # print(class_name)
class_names
['medium', 'high', 'low']
from sklearn.metrics import confusion_matrix
y_true = predictions.select("label")
y_true = y_true.toPandas()
y_pred = predictions.select("predictedLabel")
y_pred = y_pred.toPandas()
cnf_matrix = confusion_matrix(y_true, y_pred,labels=class_names)
cnf_matrix
array([[526, 11, 2],
[ 73, 33, 0],
[ 38, 0, 1]])
# Plot non-normalized confusion matrix
plt.figure()
plot_confusion_matrix(cnf_matrix, classes=class_names,
title='Confusion matrix, without normalization')
plt.show()
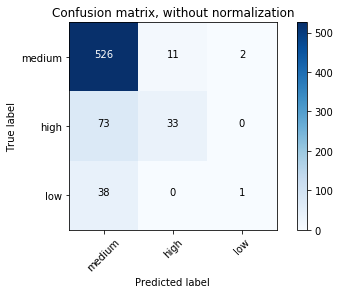
Confusion matrix, without normalization
[[526 11 2]
[ 73 33 0]
[ 38 0 1]]
# Plot normalized confusion matrix
plt.figure()
plot_confusion_matrix(cnf_matrix, classes=class_names, normalize=True,
title='Normalized confusion matrix')
plt.show()
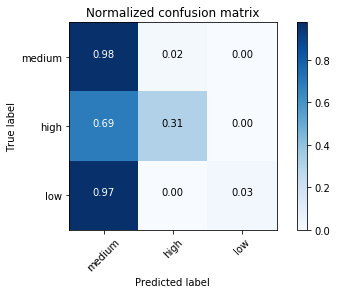
Normalized confusion matrix
[[0.97588126 0.02040816 0.00371058]
[0.68867925 0.31132075 0. ]
[0.97435897 0. 0.02564103]]
11.3. Decision tree Classification¶
11.3.1. Introduction¶
11.3.2. Demo¶
The Jupyter notebook can be download from Decision Tree Classification.
For more details, please visit DecisionTreeClassifier API .
Set up spark context and SparkSession
from pyspark.sql import SparkSession
spark = SparkSession \
.builder \
.appName("Python Spark Decision Tree classification") \
.config("spark.some.config.option", "some-value") \
.getOrCreate()
Load dataset
df = spark.read.format('com.databricks.spark.csv').\
options(header='true', \
inferschema='true') \
.load("../data/WineData2.csv",header=True);
df.show(5,True)
+-----+--------+------+-----+---------+----+-----+-------+----+---------+-------+-------+
|fixed|volatile|citric|sugar|chlorides|free|total|density| pH|sulphates|alcohol|quality|
+-----+--------+------+-----+---------+----+-----+-------+----+---------+-------+-------+
| 7.4| 0.7| 0.0| 1.9| 0.076|11.0| 34.0| 0.9978|3.51| 0.56| 9.4| 5|
| 7.8| 0.88| 0.0| 2.6| 0.098|25.0| 67.0| 0.9968| 3.2| 0.68| 9.8| 5|
| 7.8| 0.76| 0.04| 2.3| 0.092|15.0| 54.0| 0.997|3.26| 0.65| 9.8| 5|
| 11.2| 0.28| 0.56| 1.9| 0.075|17.0| 60.0| 0.998|3.16| 0.58| 9.8| 6|
| 7.4| 0.7| 0.0| 1.9| 0.076|11.0| 34.0| 0.9978|3.51| 0.56| 9.4| 5|
+-----+--------+------+-----+---------+----+-----+-------+----+---------+-------+-------+
only showing top 5 rows
# Convert to float format
def string_to_float(x):
return float(x)
#
def condition(r):
if (0<= r <= 4):
label = "low"
elif(4< r <= 6):
label = "medium"
else:
label = "high"
return label
from pyspark.sql.functions import udf
from pyspark.sql.types import StringType, DoubleType
string_to_float_udf = udf(string_to_float, DoubleType())
quality_udf = udf(lambda x: condition(x), StringType())
df = df.withColumn("quality", quality_udf("quality"))
df.show(5,True)
df.printSchema()
+-----+--------+------+-----+---------+----+-----+-------+----+---------+-------+-------+
|fixed|volatile|citric|sugar|chlorides|free|total|density| pH|sulphates|alcohol|quality|
+-----+--------+------+-----+---------+----+-----+-------+----+---------+-------+-------+
| 7.4| 0.7| 0.0| 1.9| 0.076|11.0| 34.0| 0.9978|3.51| 0.56| 9.4| medium|
| 7.8| 0.88| 0.0| 2.6| 0.098|25.0| 67.0| 0.9968| 3.2| 0.68| 9.8| medium|
| 7.8| 0.76| 0.04| 2.3| 0.092|15.0| 54.0| 0.997|3.26| 0.65| 9.8| medium|
| 11.2| 0.28| 0.56| 1.9| 0.075|17.0| 60.0| 0.998|3.16| 0.58| 9.8| medium|
| 7.4| 0.7| 0.0| 1.9| 0.076|11.0| 34.0| 0.9978|3.51| 0.56| 9.4| medium|
+-----+--------+------+-----+---------+----+-----+-------+----+---------+-------+-------+
only showing top 5 rows
root
|-- fixed: double (nullable = true)
|-- volatile: double (nullable = true)
|-- citric: double (nullable = true)
|-- sugar: double (nullable = true)
|-- chlorides: double (nullable = true)
|-- free: double (nullable = true)
|-- total: double (nullable = true)
|-- density: double (nullable = true)
|-- pH: double (nullable = true)
|-- sulphates: double (nullable = true)
|-- alcohol: double (nullable = true)
|-- quality: string (nullable = true)
Convert the data to dense vector
Note
You are strongly encouraged to try my
get_dummyfunction for dealing with the categorical data in complex dataset.Supervised learning version:
def get_dummy(df,indexCol,categoricalCols,continuousCols,labelCol): from pyspark.ml import Pipeline from pyspark.ml.feature import StringIndexer, OneHotEncoder, VectorAssembler from pyspark.sql.functions import col indexers = [ StringIndexer(inputCol=c, outputCol="{0}_indexed".format(c)) for c in categoricalCols ] # default setting: dropLast=True encoders = [ OneHotEncoder(inputCol=indexer.getOutputCol(), outputCol="{0}_encoded".format(indexer.getOutputCol())) for indexer in indexers ] assembler = VectorAssembler(inputCols=[encoder.getOutputCol() for encoder in encoders] + continuousCols, outputCol="features") pipeline = Pipeline(stages=indexers + encoders + [assembler]) model=pipeline.fit(df) data = model.transform(df) data = data.withColumn('label',col(labelCol)) return data.select(indexCol,'features','label')Unsupervised learning version:
def get_dummy(df,indexCol,categoricalCols,continuousCols): ''' Get dummy variables and concat with continuous variables for unsupervised learning. :param df: the dataframe :param categoricalCols: the name list of the categorical data :param continuousCols: the name list of the numerical data :return k: feature matrix :author: Wenqiang Feng :email: von198@gmail.com ''' indexers = [ StringIndexer(inputCol=c, outputCol="{0}_indexed".format(c)) for c in categoricalCols ] # default setting: dropLast=True encoders = [ OneHotEncoder(inputCol=indexer.getOutputCol(), outputCol="{0}_encoded".format(indexer.getOutputCol())) for indexer in indexers ] assembler = VectorAssembler(inputCols=[encoder.getOutputCol() for encoder in encoders] + continuousCols, outputCol="features") pipeline = Pipeline(stages=indexers + encoders + [assembler]) model=pipeline.fit(df) data = model.transform(df) return data.select(indexCol,'features')
Two in one:
def get_dummy(df,indexCol,categoricalCols,continuousCols,labelCol,dropLast=False): ''' Get dummy variables and concat with continuous variables for ml modeling. :param df: the dataframe :param categoricalCols: the name list of the categorical data :param continuousCols: the name list of the numerical data :param labelCol: the name of label column :param dropLast: the flag of drop last column :return: feature matrix :author: Wenqiang Feng :email: von198@gmail.com >>> df = spark.createDataFrame([ (0, "a"), (1, "b"), (2, "c"), (3, "a"), (4, "a"), (5, "c") ], ["id", "category"]) >>> indexCol = 'id' >>> categoricalCols = ['category'] >>> continuousCols = [] >>> labelCol = [] >>> mat = get_dummy(df,indexCol,categoricalCols,continuousCols,labelCol) >>> mat.show() >>> +---+-------------+ | id| features| +---+-------------+ | 0|[1.0,0.0,0.0]| | 1|[0.0,0.0,1.0]| | 2|[0.0,1.0,0.0]| | 3|[1.0,0.0,0.0]| | 4|[1.0,0.0,0.0]| | 5|[0.0,1.0,0.0]| +---+-------------+ ''' from pyspark.ml import Pipeline from pyspark.ml.feature import StringIndexer, OneHotEncoder, VectorAssembler from pyspark.sql.functions import col indexers = [ StringIndexer(inputCol=c, outputCol="{0}_indexed".format(c)) for c in categoricalCols ] # default setting: dropLast=True encoders = [ OneHotEncoder(inputCol=indexer.getOutputCol(), outputCol="{0}_encoded".format(indexer.getOutputCol()),dropLast=dropLast) for indexer in indexers ] assembler = VectorAssembler(inputCols=[encoder.getOutputCol() for encoder in encoders] + continuousCols, outputCol="features") pipeline = Pipeline(stages=indexers + encoders + [assembler]) model=pipeline.fit(df) data = model.transform(df) if indexCol and labelCol: # for supervised learning data = data.withColumn('label',col(labelCol)) return data.select(indexCol,'features','label') elif not indexCol and labelCol: # for supervised learning data = data.withColumn('label',col(labelCol)) return data.select('features','label') elif indexCol and not labelCol: # for unsupervised learning return data.select(indexCol,'features') elif not indexCol and not labelCol: # for unsupervised learning return data.select('features')
# !!!!caution: not from pyspark.mllib.linalg import Vectors
from pyspark.ml.linalg import Vectors
from pyspark.ml import Pipeline
from pyspark.ml.feature import IndexToString,StringIndexer, VectorIndexer
from pyspark.ml.tuning import CrossValidator, ParamGridBuilder
from pyspark.ml.evaluation import MulticlassClassificationEvaluator
def transData(data):
return data.rdd.map(lambda r: [Vectors.dense(r[:-1]),r[-1]]).toDF(['features','label'])
Transform the dataset to DataFrame
transformed = transData(df)
transformed.show(5)
+--------------------+------+
| features| label|
+--------------------+------+
|[7.4,0.7,0.0,1.9,...|medium|
|[7.8,0.88,0.0,2.6...|medium|
|[7.8,0.76,0.04,2....|medium|
|[11.2,0.28,0.56,1...|medium|
|[7.4,0.7,0.0,1.9,...|medium|
+--------------------+------+
only showing top 5 rows
Deal with Categorical Label and Variables
# Index labels, adding metadata to the label column
labelIndexer = StringIndexer(inputCol='label',
outputCol='indexedLabel').fit(transformed)
labelIndexer.transform(transformed).show(5, True)
+--------------------+------+------------+
| features| label|indexedLabel|
+--------------------+------+------------+
|[7.4,0.7,0.0,1.9,...|medium| 0.0|
|[7.8,0.88,0.0,2.6...|medium| 0.0|
|[7.8,0.76,0.04,2....|medium| 0.0|
|[11.2,0.28,0.56,1...|medium| 0.0|
|[7.4,0.7,0.0,1.9,...|medium| 0.0|
+--------------------+------+------------+
only showing top 5 rows
# Automatically identify categorical features, and index them.
# Set maxCategories so features with > 4 distinct values are treated as continuous.
featureIndexer =VectorIndexer(inputCol="features", \
outputCol="indexedFeatures", \
maxCategories=4).fit(transformed)
featureIndexer.transform(transformed).show(5, True)
+--------------------+------+--------------------+
| features| label| indexedFeatures|
+--------------------+------+--------------------+
|[7.4,0.7,0.0,1.9,...|medium|[7.4,0.7,0.0,1.9,...|
|[7.8,0.88,0.0,2.6...|medium|[7.8,0.88,0.0,2.6...|
|[7.8,0.76,0.04,2....|medium|[7.8,0.76,0.04,2....|
|[11.2,0.28,0.56,1...|medium|[11.2,0.28,0.56,1...|
|[7.4,0.7,0.0,1.9,...|medium|[7.4,0.7,0.0,1.9,...|
+--------------------+------+--------------------+
only showing top 5 rows
Split the data to training and test data sets
# Split the data into training and test sets (40% held out for testing)
(trainingData, testData) = transformed.randomSplit([0.6, 0.4])
trainingData.show(5)
testData.show(5)
+--------------------+------+
| features| label|
+--------------------+------+
|[4.6,0.52,0.15,2....| low|
|[4.7,0.6,0.17,2.3...|medium|
|[5.0,1.02,0.04,1....| low|
|[5.0,1.04,0.24,1....|medium|
|[5.1,0.585,0.0,1....| high|
+--------------------+------+
only showing top 5 rows
+--------------------+------+
| features| label|
+--------------------+------+
|[4.9,0.42,0.0,2.1...| high|
|[5.0,0.38,0.01,1....|medium|
|[5.0,0.4,0.5,4.3,...|medium|
|[5.0,0.42,0.24,2....| high|
|[5.0,0.74,0.0,1.2...|medium|
+--------------------+------+
only showing top 5 rows
Fit Decision Tree Classification Model
from pyspark.ml.classification import DecisionTreeClassifier
# Train a DecisionTree model
dTree = DecisionTreeClassifier(labelCol='indexedLabel', featuresCol='indexedFeatures')
Pipeline Architecture
# Convert indexed labels back to original labels.
labelConverter = IndexToString(inputCol="prediction", outputCol="predictedLabel",
labels=labelIndexer.labels)
# Chain indexers and tree in a Pipeline
pipeline = Pipeline(stages=[labelIndexer, featureIndexer, dTree,labelConverter])
# Train model. This also runs the indexers.
model = pipeline.fit(trainingData)
Make predictions
# Make predictions.
predictions = model.transform(testData)
# Select example rows to display.
predictions.select("features","label","predictedLabel").show(5)
+--------------------+------+--------------+
| features| label|predictedLabel|
+--------------------+------+--------------+
|[4.9,0.42,0.0,2.1...| high| high|
|[5.0,0.38,0.01,1....|medium| medium|
|[5.0,0.4,0.5,4.3,...|medium| medium|
|[5.0,0.42,0.24,2....| high| medium|
|[5.0,0.74,0.0,1.2...|medium| medium|
+--------------------+------+--------------+
only showing top 5 rows
Evaluation
from pyspark.ml.evaluation import MulticlassClassificationEvaluator
# Select (prediction, true label) and compute test error
evaluator = MulticlassClassificationEvaluator(
labelCol="indexedLabel", predictionCol="prediction", metricName="accuracy")
accuracy = evaluator.evaluate(predictions)
print("Test Error = %g" % (1.0 - accuracy))
rfModel = model.stages[-2]
print(rfModel) # summary only
Test Error = 0.45509
DecisionTreeClassificationModel (uid=DecisionTreeClassifier_4545ac8dca9c8438ef2a)
of depth 5 with 59 nodes
visualization
import matplotlib.pyplot as plt
import numpy as np
import itertools
def plot_confusion_matrix(cm, classes,
normalize=False,
title='Confusion matrix',
cmap=plt.cm.Blues):
"""
This function prints and plots the confusion matrix.
Normalization can be applied by setting `normalize=True`.
"""
if normalize:
cm = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]
print("Normalized confusion matrix")
else:
print('Confusion matrix, without normalization')
print(cm)
plt.imshow(cm, interpolation='nearest', cmap=cmap)
plt.title(title)
plt.colorbar()
tick_marks = np.arange(len(classes))
plt.xticks(tick_marks, classes, rotation=45)
plt.yticks(tick_marks, classes)
fmt = '.2f' if normalize else 'd'
thresh = cm.max() / 2.
for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):
plt.text(j, i, format(cm[i, j], fmt),
horizontalalignment="center",
color="white" if cm[i, j] > thresh else "black")
plt.tight_layout()
plt.ylabel('True label')
plt.xlabel('Predicted label')
class_temp = predictions.select("label").groupBy("label")\
.count().sort('count', ascending=False).toPandas()
class_temp = class_temp["label"].values.tolist()
class_names = map(str, class_temp)
# # # print(class_name)
class_names
['medium', 'high', 'low']
from sklearn.metrics import confusion_matrix
y_true = predictions.select("label")
y_true = y_true.toPandas()
y_pred = predictions.select("predictedLabel")
y_pred = y_pred.toPandas()
cnf_matrix = confusion_matrix(y_true, y_pred,labels=class_names)
cnf_matrix
array([[497, 29, 7],
[ 40, 42, 0],
[ 22, 0, 2]])
# Plot non-normalized confusion matrix
plt.figure()
plot_confusion_matrix(cnf_matrix, classes=class_names,
title='Confusion matrix, without normalization')
plt.show()
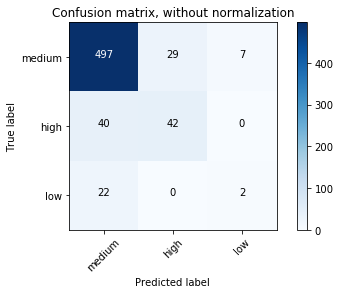
Confusion matrix, without normalization
[[497 29 7]
[ 40 42 0]
[ 22 0 2]]
# Plot normalized confusion matrix
plt.figure()
plot_confusion_matrix(cnf_matrix, classes=class_names, normalize=True,
title='Normalized confusion matrix')
plt.show()
Normalized confusion matrix
[[ 0.93245779 0.05440901 0.01313321]
[ 0.48780488 0.51219512 0. ]
[ 0.91666667 0. 0.08333333]]

11.4. Random forest Classification¶
11.4.1. Introduction¶
11.4.2. Demo¶
The Jupyter notebook can be download from Random forest Classification.
For more details, please visit RandomForestClassifier API .
Set up spark context and SparkSession
from pyspark.sql import SparkSession
spark = SparkSession \
.builder \
.appName("Python Spark Decision Tree classification") \
.config("spark.some.config.option", "some-value") \
.getOrCreate()
Load dataset
df = spark.read.format('com.databricks.spark.csv').\
options(header='true', \
inferschema='true') \
.load("../data/WineData2.csv",header=True);
df.show(5,True)
+-----+--------+------+-----+---------+----+-----+-------+----+---------+-------+-------+
|fixed|volatile|citric|sugar|chlorides|free|total|density| pH|sulphates|alcohol|quality|
+-----+--------+------+-----+---------+----+-----+-------+----+---------+-------+-------+
| 7.4| 0.7| 0.0| 1.9| 0.076|11.0| 34.0| 0.9978|3.51| 0.56| 9.4| 5|
| 7.8| 0.88| 0.0| 2.6| 0.098|25.0| 67.0| 0.9968| 3.2| 0.68| 9.8| 5|
| 7.8| 0.76| 0.04| 2.3| 0.092|15.0| 54.0| 0.997|3.26| 0.65| 9.8| 5|
| 11.2| 0.28| 0.56| 1.9| 0.075|17.0| 60.0| 0.998|3.16| 0.58| 9.8| 6|
| 7.4| 0.7| 0.0| 1.9| 0.076|11.0| 34.0| 0.9978|3.51| 0.56| 9.4| 5|
+-----+--------+------+-----+---------+----+-----+-------+----+---------+-------+-------+
only showing top 5 rows
# Convert to float format
def string_to_float(x):
return float(x)
#
def condition(r):
if (0<= r <= 4):
label = "low"
elif(4< r <= 6):
label = "medium"
else:
label = "high"
return label
from pyspark.sql.functions import udf
from pyspark.sql.types import StringType, DoubleType
string_to_float_udf = udf(string_to_float, DoubleType())
quality_udf = udf(lambda x: condition(x), StringType())
df = df.withColumn("quality", quality_udf("quality"))
df.show(5,True)
df.printSchema()
+-----+--------+------+-----+---------+----+-----+-------+----+---------+-------+-------+
|fixed|volatile|citric|sugar|chlorides|free|total|density| pH|sulphates|alcohol|quality|
+-----+--------+------+-----+---------+----+-----+-------+----+---------+-------+-------+
| 7.4| 0.7| 0.0| 1.9| 0.076|11.0| 34.0| 0.9978|3.51| 0.56| 9.4| medium|
| 7.8| 0.88| 0.0| 2.6| 0.098|25.0| 67.0| 0.9968| 3.2| 0.68| 9.8| medium|
| 7.8| 0.76| 0.04| 2.3| 0.092|15.0| 54.0| 0.997|3.26| 0.65| 9.8| medium|
| 11.2| 0.28| 0.56| 1.9| 0.075|17.0| 60.0| 0.998|3.16| 0.58| 9.8| medium|
| 7.4| 0.7| 0.0| 1.9| 0.076|11.0| 34.0| 0.9978|3.51| 0.56| 9.4| medium|
+-----+--------+------+-----+---------+----+-----+-------+----+---------+-------+-------+
only showing top 5 rows
root
|-- fixed: double (nullable = true)
|-- volatile: double (nullable = true)
|-- citric: double (nullable = true)
|-- sugar: double (nullable = true)
|-- chlorides: double (nullable = true)
|-- free: double (nullable = true)
|-- total: double (nullable = true)
|-- density: double (nullable = true)
|-- pH: double (nullable = true)
|-- sulphates: double (nullable = true)
|-- alcohol: double (nullable = true)
|-- quality: string (nullable = true)
Convert the data to dense vector
Note
You are strongly encouraged to try my
get_dummyfunction for dealing with the categorical data in complex dataset.Supervised learning version:
def get_dummy(df,indexCol,categoricalCols,continuousCols,labelCol): from pyspark.ml import Pipeline from pyspark.ml.feature import StringIndexer, OneHotEncoder, VectorAssembler from pyspark.sql.functions import col indexers = [ StringIndexer(inputCol=c, outputCol="{0}_indexed".format(c)) for c in categoricalCols ] # default setting: dropLast=True encoders = [ OneHotEncoder(inputCol=indexer.getOutputCol(), outputCol="{0}_encoded".format(indexer.getOutputCol())) for indexer in indexers ] assembler = VectorAssembler(inputCols=[encoder.getOutputCol() for encoder in encoders] + continuousCols, outputCol="features") pipeline = Pipeline(stages=indexers + encoders + [assembler]) model=pipeline.fit(df) data = model.transform(df) data = data.withColumn('label',col(labelCol)) return data.select(indexCol,'features','label')Unsupervised learning version:
def get_dummy(df,indexCol,categoricalCols,continuousCols): ''' Get dummy variables and concat with continuous variables for unsupervised learning. :param df: the dataframe :param categoricalCols: the name list of the categorical data :param continuousCols: the name list of the numerical data :return k: feature matrix :author: Wenqiang Feng :email: von198@gmail.com ''' indexers = [ StringIndexer(inputCol=c, outputCol="{0}_indexed".format(c)) for c in categoricalCols ] # default setting: dropLast=True encoders = [ OneHotEncoder(inputCol=indexer.getOutputCol(), outputCol="{0}_encoded".format(indexer.getOutputCol())) for indexer in indexers ] assembler = VectorAssembler(inputCols=[encoder.getOutputCol() for encoder in encoders] + continuousCols, outputCol="features") pipeline = Pipeline(stages=indexers + encoders + [assembler]) model=pipeline.fit(df) data = model.transform(df) return data.select(indexCol,'features')
Two in one:
def get_dummy(df,indexCol,categoricalCols,continuousCols,labelCol,dropLast=False): ''' Get dummy variables and concat with continuous variables for ml modeling. :param df: the dataframe :param categoricalCols: the name list of the categorical data :param continuousCols: the name list of the numerical data :param labelCol: the name of label column :param dropLast: the flag of drop last column :return: feature matrix :author: Wenqiang Feng :email: von198@gmail.com >>> df = spark.createDataFrame([ (0, "a"), (1, "b"), (2, "c"), (3, "a"), (4, "a"), (5, "c") ], ["id", "category"]) >>> indexCol = 'id' >>> categoricalCols = ['category'] >>> continuousCols = [] >>> labelCol = [] >>> mat = get_dummy(df,indexCol,categoricalCols,continuousCols,labelCol) >>> mat.show() >>> +---+-------------+ | id| features| +---+-------------+ | 0|[1.0,0.0,0.0]| | 1|[0.0,0.0,1.0]| | 2|[0.0,1.0,0.0]| | 3|[1.0,0.0,0.0]| | 4|[1.0,0.0,0.0]| | 5|[0.0,1.0,0.0]| +---+-------------+ ''' from pyspark.ml import Pipeline from pyspark.ml.feature import StringIndexer, OneHotEncoder, VectorAssembler from pyspark.sql.functions import col indexers = [ StringIndexer(inputCol=c, outputCol="{0}_indexed".format(c)) for c in categoricalCols ] # default setting: dropLast=True encoders = [ OneHotEncoder(inputCol=indexer.getOutputCol(), outputCol="{0}_encoded".format(indexer.getOutputCol()),dropLast=dropLast) for indexer in indexers ] assembler = VectorAssembler(inputCols=[encoder.getOutputCol() for encoder in encoders] + continuousCols, outputCol="features") pipeline = Pipeline(stages=indexers + encoders + [assembler]) model=pipeline.fit(df) data = model.transform(df) if indexCol and labelCol: # for supervised learning data = data.withColumn('label',col(labelCol)) return data.select(indexCol,'features','label') elif not indexCol and labelCol: # for supervised learning data = data.withColumn('label',col(labelCol)) return data.select('features','label') elif indexCol and not labelCol: # for unsupervised learning return data.select(indexCol,'features') elif not indexCol and not labelCol: # for unsupervised learning return data.select('features')
# !!!!caution: not from pyspark.mllib.linalg import Vectors
from pyspark.ml.linalg import Vectors
from pyspark.ml import Pipeline
from pyspark.ml.feature import IndexToString,StringIndexer, VectorIndexer
from pyspark.ml.tuning import CrossValidator, ParamGridBuilder
from pyspark.ml.evaluation import MulticlassClassificationEvaluator
def transData(data):
return data.rdd.map(lambda r: [Vectors.dense(r[:-1]),r[-1]]).toDF(['features','label'])
Transform the dataset to DataFrame
transformed = transData(df)
transformed.show(5)
+--------------------+------+
| features| label|
+--------------------+------+
|[7.4,0.7,0.0,1.9,...|medium|
|[7.8,0.88,0.0,2.6...|medium|
|[7.8,0.76,0.04,2....|medium|
|[11.2,0.28,0.56,1...|medium|
|[7.4,0.7,0.0,1.9,...|medium|
+--------------------+------+
only showing top 5 rows
Deal with Categorical Label and Variables
# Index labels, adding metadata to the label column
labelIndexer = StringIndexer(inputCol='label',
outputCol='indexedLabel').fit(transformed)
labelIndexer.transform(transformed).show(5, True)
+--------------------+------+------------+
| features| label|indexedLabel|
+--------------------+------+------------+
|[7.4,0.7,0.0,1.9,...|medium| 0.0|
|[7.8,0.88,0.0,2.6...|medium| 0.0|
|[7.8,0.76,0.04,2....|medium| 0.0|
|[11.2,0.28,0.56,1...|medium| 0.0|
|[7.4,0.7,0.0,1.9,...|medium| 0.0|
+--------------------+------+------------+
only showing top 5 rows
# Automatically identify categorical features, and index them.
# Set maxCategories so features with > 4 distinct values are treated as continuous.
featureIndexer =VectorIndexer(inputCol="features", \
outputCol="indexedFeatures", \
maxCategories=4).fit(transformed)
featureIndexer.transform(transformed).show(5, True)
+--------------------+------+--------------------+
| features| label| indexedFeatures|
+--------------------+------+--------------------+
|[7.4,0.7,0.0,1.9,...|medium|[7.4,0.7,0.0,1.9,...|
|[7.8,0.88,0.0,2.6...|medium|[7.8,0.88,0.0,2.6...|
|[7.8,0.76,0.04,2....|medium|[7.8,0.76,0.04,2....|
|[11.2,0.28,0.56,1...|medium|[11.2,0.28,0.56,1...|
|[7.4,0.7,0.0,1.9,...|medium|[7.4,0.7,0.0,1.9,...|
+--------------------+------+--------------------+
only showing top 5 rows
Split the data to training and test data sets
# Split the data into training and test sets (40% held out for testing)
(trainingData, testData) = transformed.randomSplit([0.6, 0.4])
trainingData.show(5)
testData.show(5)
+--------------------+------+
| features| label|
+--------------------+------+
|[4.6,0.52,0.15,2....| low|
|[4.7,0.6,0.17,2.3...|medium|
|[5.0,1.02,0.04,1....| low|
|[5.0,1.04,0.24,1....|medium|
|[5.1,0.585,0.0,1....| high|
+--------------------+------+
only showing top 5 rows
+--------------------+------+
| features| label|
+--------------------+------+
|[4.9,0.42,0.0,2.1...| high|
|[5.0,0.38,0.01,1....|medium|
|[5.0,0.4,0.5,4.3,...|medium|
|[5.0,0.42,0.24,2....| high|
|[5.0,0.74,0.0,1.2...|medium|
+--------------------+------+
only showing top 5 rows
Fit Random Forest Classification Model
from pyspark.ml.classification import RandomForestClassifier
# Train a RandomForest model.
rf = RandomForestClassifier(labelCol="indexedLabel", featuresCol="indexedFeatures", numTrees=10)
Pipeline Architecture
# Convert indexed labels back to original labels.
labelConverter = IndexToString(inputCol="prediction", outputCol="predictedLabel",
labels=labelIndexer.labels)
# Chain indexers and tree in a Pipeline
pipeline = Pipeline(stages=[labelIndexer, featureIndexer, rf,labelConverter])
# Train model. This also runs the indexers.
model = pipeline.fit(trainingData)
Make predictions
# Make predictions.
predictions = model.transform(testData)
# Select example rows to display.
predictions.select("features","label","predictedLabel").show(5)
+--------------------+------+--------------+
| features| label|predictedLabel|
+--------------------+------+--------------+
|[4.9,0.42,0.0,2.1...| high| high|
|[5.0,0.38,0.01,1....|medium| medium|
|[5.0,0.4,0.5,4.3,...|medium| medium|
|[5.0,0.42,0.24,2....| high| medium|
|[5.0,0.74,0.0,1.2...|medium| medium|
+--------------------+------+--------------+
only showing top 5 rows
Evaluation
from pyspark.ml.evaluation import MulticlassClassificationEvaluator
# Select (prediction, true label) and compute test error
evaluator = MulticlassClassificationEvaluator(
labelCol="indexedLabel", predictionCol="prediction", metricName="accuracy")
accuracy = evaluator.evaluate(predictions)
print("Test Error = %g" % (1.0 - accuracy))
rfModel = model.stages[-2]
print(rfModel) # summary only
Test Error = 0.173502
RandomForestClassificationModel (uid=rfc_a3395531f1d2) with 10 trees
visualization
import matplotlib.pyplot as plt
import numpy as np
import itertools
def plot_confusion_matrix(cm, classes,
normalize=False,
title='Confusion matrix',
cmap=plt.cm.Blues):
"""
This function prints and plots the confusion matrix.
Normalization can be applied by setting `normalize=True`.
"""
if normalize:
cm = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]
print("Normalized confusion matrix")
else:
print('Confusion matrix, without normalization')
print(cm)
plt.imshow(cm, interpolation='nearest', cmap=cmap)
plt.title(title)
plt.colorbar()
tick_marks = np.arange(len(classes))
plt.xticks(tick_marks, classes, rotation=45)
plt.yticks(tick_marks, classes)
fmt = '.2f' if normalize else 'd'
thresh = cm.max() / 2.
for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):
plt.text(j, i, format(cm[i, j], fmt),
horizontalalignment="center",
color="white" if cm[i, j] > thresh else "black")
plt.tight_layout()
plt.ylabel('True label')
plt.xlabel('Predicted label')
class_temp = predictions.select("label").groupBy("label")\
.count().sort('count', ascending=False).toPandas()
class_temp = class_temp["label"].values.tolist()
class_names = map(str, class_temp)
# # # print(class_name)
class_names
['medium', 'high', 'low']
from sklearn.metrics import confusion_matrix
y_true = predictions.select("label")
y_true = y_true.toPandas()
y_pred = predictions.select("predictedLabel")
y_pred = y_pred.toPandas()
cnf_matrix = confusion_matrix(y_true, y_pred,labels=class_names)
cnf_matrix
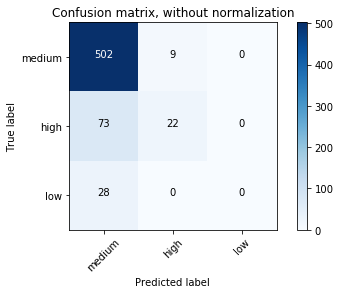
array([[502, 9, 0],
[ 73, 22, 0],
[ 28, 0, 0]])
# Plot non-normalized confusion matrix
plt.figure()
plot_confusion_matrix(cnf_matrix, classes=class_names,
title='Confusion matrix, without normalization')
plt.show()
Confusion matrix, without normalization
[[502 9 0]
[ 73 22 0]
[ 28 0 0]]
# Plot normalized confusion matrix
plt.figure()
plot_confusion_matrix(cnf_matrix, classes=class_names, normalize=True,
title='Normalized confusion matrix')
plt.show()
Normalized confusion matrix
[[ 0.98238748 0.01761252 0. ]
[ 0.76842105 0.23157895 0. ]
[ 1. 0. 0. ]]

11.5. Gradient-boosted tree Classification¶
11.5.1. Introduction¶
11.5.2. Demo¶
The Jupyter notebook can be download from Gradient boosted tree Classification.
For more details, please visit GBTClassifier API .
Warning
Unfortunately, the GBTClassifier currently only supports binary labels.
11.6. XGBoost: Gradient-boosted tree Classification¶
11.6.1. Introduction¶
11.6.2. Demo¶
The Jupyter notebook can be download from Gradient boosted tree Classification.
For more details, please visit GBTClassifier API .
Warning
Unfortunately, I didn’t find a good way to setup the XGBoost directly in Spark. But I do get the XGBoost work with pysparkling
on my machine.
Start H2O cluster inside the Spark environment
from pysparkling import *
hc = H2OContext.getOrCreate(spark)
Connecting to H2O server at http://192.168.0.102:54323... successful.
H2O cluster uptime: 07 secs
H2O cluster timezone: America/Chicago
H2O data parsing timezone: UTC
H2O cluster version: 3.22.1.3
H2O cluster version age: 20 days
H2O cluster name: sparkling-water-dt216661_local-1550259209801
H2O cluster total nodes: 1
H2O cluster free memory: 848 Mb
H2O cluster total cores: 8
H2O cluster allowed cores: 8
H2O cluster status: accepting new members, healthy
H2O connection url: http://192.168.0.102:54323
H2O connection proxy: None
H2O internal security: False
H2O API Extensions: XGBoost, Algos, AutoML, Core V3, Core V4
Python version: 3.7.1 final
Sparkling Water Context:
* H2O name: sparkling-water-dt216661_local-1550259209801
* cluster size: 1
* list of used nodes:
(executorId, host, port)
------------------------
(driver,192.168.0.102,54323)
------------------------
Open H2O Flow in browser: http://192.168.0.102:54323 (CMD + click in Mac OSX)
Parse the data using H2O and convert them to Spark Frame
import h2o
frame = h2o.import_file("https://raw.githubusercontent.com/h2oai/sparkling-water/master/examples/smalldata/prostate/prostate.csv")
spark_frame = hc.as_spark_frame(frame)

spark_frame.show(4)
+---+-------+---+----+-----+-----+----+----+-------+
| ID|CAPSULE|AGE|RACE|DPROS|DCAPS| PSA| VOL|GLEASON|
+---+-------+---+----+-----+-----+----+----+-------+
| 1| 0| 65| 1| 2| 1| 1.4| 0.0| 6|
| 2| 0| 72| 1| 3| 2| 6.7| 0.0| 7|
| 3| 0| 70| 1| 1| 2| 4.9| 0.0| 6|
| 4| 0| 76| 2| 2| 1|51.2|20.0| 7|
+---+-------+---+----+-----+-----+----+----+-------+
only showing top 4 rows
Train the model
from pysparkling.ml import H2OXGBoost
estimator = H2OXGBoost(predictionCol="AGE")
model = estimator.fit(spark_frame)
Run Predictions
predictions = model.transform(spark_frame)
predictions.show(4)
+---+-------+---+----+-----+-----+----+----+-------+-------------------+
| ID|CAPSULE|AGE|RACE|DPROS|DCAPS| PSA| VOL|GLEASON| prediction_output|
+---+-------+---+----+-----+-----+----+----+-------+-------------------+
| 1| 0| 65| 1| 2| 1| 1.4| 0.0| 6|[64.85852813720703]|
| 2| 0| 72| 1| 3| 2| 6.7| 0.0| 7| [72.0611801147461]|
| 3| 0| 70| 1| 1| 2| 4.9| 0.0| 6|[70.26496887207031]|
| 4| 0| 76| 2| 2| 1|51.2|20.0| 7|[75.26521301269531]|
+---+-------+---+----+-----+-----+----+----+-------+-------------------+
only showing top 4 rows
11.7. Naive Bayes Classification¶
11.7.1. Introduction¶
11.7.2. Demo¶
The Jupyter notebook can be download from Naive Bayes Classification.
For more details, please visit NaiveBayes API .
Set up spark context and SparkSession
from pyspark.sql import SparkSession
spark = SparkSession \
.builder \
.appName("Python Spark Naive Bayes classification") \
.config("spark.some.config.option", "some-value") \
.getOrCreate()
Load dataset
df = spark.read.format('com.databricks.spark.csv') \
.options(header='true', inferschema='true') \
.load("./data/WineData2.csv",header=True);
df.show(5)
+-----+--------+------+-----+---------+----+-----+-------+----+---------+-------+-------+
|fixed|volatile|citric|sugar|chlorides|free|total|density| pH|sulphates|alcohol|quality|
+-----+--------+------+-----+---------+----+-----+-------+----+---------+-------+-------+
| 7.4| 0.7| 0.0| 1.9| 0.076|11.0| 34.0| 0.9978|3.51| 0.56| 9.4| 5|
| 7.8| 0.88| 0.0| 2.6| 0.098|25.0| 67.0| 0.9968| 3.2| 0.68| 9.8| 5|
| 7.8| 0.76| 0.04| 2.3| 0.092|15.0| 54.0| 0.997|3.26| 0.65| 9.8| 5|
| 11.2| 0.28| 0.56| 1.9| 0.075|17.0| 60.0| 0.998|3.16| 0.58| 9.8| 6|
| 7.4| 0.7| 0.0| 1.9| 0.076|11.0| 34.0| 0.9978|3.51| 0.56| 9.4| 5|
+-----+--------+------+-----+---------+----+-----+-------+----+---------+-------+-------+
only showing top 5 rows
df.printSchema()
root
|-- fixed: double (nullable = true)
|-- volatile: double (nullable = true)
|-- citric: double (nullable = true)
|-- sugar: double (nullable = true)
|-- chlorides: double (nullable = true)
|-- free: double (nullable = true)
|-- total: double (nullable = true)
|-- density: double (nullable = true)
|-- pH: double (nullable = true)
|-- sulphates: double (nullable = true)
|-- alcohol: double (nullable = true)
|-- quality: string (nullable = true)
# Convert to float format
def string_to_float(x):
return float(x)
#
def condition(r):
if (0<= r <= 6):
label = "low"
else:
label = "high"
return label
from pyspark.sql.functions import udf
from pyspark.sql.types import StringType, DoubleType
string_to_float_udf = udf(string_to_float, DoubleType())
quality_udf = udf(lambda x: condition(x), StringType())
df = df.withColumn("quality", quality_udf("quality"))
df.show(5,True)
+-----+--------+------+-----+---------+----+-----+-------+----+---------+-------+-------+
|fixed|volatile|citric|sugar|chlorides|free|total|density| pH|sulphates|alcohol|quality|
+-----+--------+------+-----+---------+----+-----+-------+----+---------+-------+-------+
| 7.4| 0.7| 0.0| 1.9| 0.076|11.0| 34.0| 0.9978|3.51| 0.56| 9.4| medium|
| 7.8| 0.88| 0.0| 2.6| 0.098|25.0| 67.0| 0.9968| 3.2| 0.68| 9.8| medium|
| 7.8| 0.76| 0.04| 2.3| 0.092|15.0| 54.0| 0.997|3.26| 0.65| 9.8| medium|
| 11.2| 0.28| 0.56| 1.9| 0.075|17.0| 60.0| 0.998|3.16| 0.58| 9.8| medium|
| 7.4| 0.7| 0.0| 1.9| 0.076|11.0| 34.0| 0.9978|3.51| 0.56| 9.4| medium|
+-----+--------+------+-----+---------+----+-----+-------+----+---------+-------+-------+
only showing top 5 rows
df.printSchema()
root
|-- fixed: double (nullable = true)
|-- volatile: double (nullable = true)
|-- citric: double (nullable = true)
|-- sugar: double (nullable = true)
|-- chlorides: double (nullable = true)
|-- free: double (nullable = true)
|-- total: double (nullable = true)
|-- density: double (nullable = true)
|-- pH: double (nullable = true)
|-- sulphates: double (nullable = true)
|-- alcohol: double (nullable = true)
|-- quality: string (nullable = true)
Deal with categorical data and Convert the data to dense vector
Note
You are strongly encouraged to try my
get_dummyfunction for dealing with the categorical data in complex dataset.Supervised learning version:
def get_dummy(df,indexCol,categoricalCols,continuousCols,labelCol): from pyspark.ml import Pipeline from pyspark.ml.feature import StringIndexer, OneHotEncoder, VectorAssembler from pyspark.sql.functions import col indexers = [ StringIndexer(inputCol=c, outputCol="{0}_indexed".format(c)) for c in categoricalCols ] # default setting: dropLast=True encoders = [ OneHotEncoder(inputCol=indexer.getOutputCol(), outputCol="{0}_encoded".format(indexer.getOutputCol())) for indexer in indexers ] assembler = VectorAssembler(inputCols=[encoder.getOutputCol() for encoder in encoders] + continuousCols, outputCol="features") pipeline = Pipeline(stages=indexers + encoders + [assembler]) model=pipeline.fit(df) data = model.transform(df) data = data.withColumn('label',col(labelCol)) return data.select(indexCol,'features','label')Unsupervised learning version:
def get_dummy(df,indexCol,categoricalCols,continuousCols): ''' Get dummy variables and concat with continuous variables for unsupervised learning. :param df: the dataframe :param categoricalCols: the name list of the categorical data :param continuousCols: the name list of the numerical data :return k: feature matrix :author: Wenqiang Feng :email: von198@gmail.com ''' indexers = [ StringIndexer(inputCol=c, outputCol="{0}_indexed".format(c)) for c in categoricalCols ] # default setting: dropLast=True encoders = [ OneHotEncoder(inputCol=indexer.getOutputCol(), outputCol="{0}_encoded".format(indexer.getOutputCol())) for indexer in indexers ] assembler = VectorAssembler(inputCols=[encoder.getOutputCol() for encoder in encoders] + continuousCols, outputCol="features") pipeline = Pipeline(stages=indexers + encoders + [assembler]) model=pipeline.fit(df) data = model.transform(df) return data.select(indexCol,'features')
Two in one:
def get_dummy(df,indexCol,categoricalCols,continuousCols,labelCol,dropLast=False): ''' Get dummy variables and concat with continuous variables for ml modeling. :param df: the dataframe :param categoricalCols: the name list of the categorical data :param continuousCols: the name list of the numerical data :param labelCol: the name of label column :param dropLast: the flag of drop last column :return: feature matrix :author: Wenqiang Feng :email: von198@gmail.com >>> df = spark.createDataFrame([ (0, "a"), (1, "b"), (2, "c"), (3, "a"), (4, "a"), (5, "c") ], ["id", "category"]) >>> indexCol = 'id' >>> categoricalCols = ['category'] >>> continuousCols = [] >>> labelCol = [] >>> mat = get_dummy(df,indexCol,categoricalCols,continuousCols,labelCol) >>> mat.show() >>> +---+-------------+ | id| features| +---+-------------+ | 0|[1.0,0.0,0.0]| | 1|[0.0,0.0,1.0]| | 2|[0.0,1.0,0.0]| | 3|[1.0,0.0,0.0]| | 4|[1.0,0.0,0.0]| | 5|[0.0,1.0,0.0]| +---+-------------+ ''' from pyspark.ml import Pipeline from pyspark.ml.feature import StringIndexer, OneHotEncoder, VectorAssembler from pyspark.sql.functions import col indexers = [ StringIndexer(inputCol=c, outputCol="{0}_indexed".format(c)) for c in categoricalCols ] # default setting: dropLast=True encoders = [ OneHotEncoder(inputCol=indexer.getOutputCol(), outputCol="{0}_encoded".format(indexer.getOutputCol()),dropLast=dropLast) for indexer in indexers ] assembler = VectorAssembler(inputCols=[encoder.getOutputCol() for encoder in encoders] + continuousCols, outputCol="features") pipeline = Pipeline(stages=indexers + encoders + [assembler]) model=pipeline.fit(df) data = model.transform(df) if indexCol and labelCol: # for supervised learning data = data.withColumn('label',col(labelCol)) return data.select(indexCol,'features','label') elif not indexCol and labelCol: # for supervised learning data = data.withColumn('label',col(labelCol)) return data.select('features','label') elif indexCol and not labelCol: # for unsupervised learning return data.select(indexCol,'features') elif not indexCol and not labelCol: # for unsupervised learning return data.select('features')
def get_dummy(df,categoricalCols,continuousCols,labelCol):
from pyspark.ml import Pipeline
from pyspark.ml.feature import StringIndexer, OneHotEncoder, VectorAssembler
from pyspark.sql.functions import col
indexers = [ StringIndexer(inputCol=c, outputCol="{0}_indexed".format(c))
for c in categoricalCols ]
# default setting: dropLast=True
encoders = [ OneHotEncoder(inputCol=indexer.getOutputCol(),
outputCol="{0}_encoded".format(indexer.getOutputCol()))
for indexer in indexers ]
assembler = VectorAssembler(inputCols=[encoder.getOutputCol() for encoder in encoders]
+ continuousCols, outputCol="features")
pipeline = Pipeline(stages=indexers + encoders + [assembler])
model=pipeline.fit(df)
data = model.transform(df)
data = data.withColumn('label',col(labelCol))
return data.select('features','label')
Transform the dataset to DataFrame
from pyspark.ml.linalg import Vectors # !!!!caution: not from pyspark.mllib.linalg import Vectors
from pyspark.ml import Pipeline
from pyspark.ml.feature import IndexToString,StringIndexer, VectorIndexer
from pyspark.ml.tuning import CrossValidator, ParamGridBuilder
from pyspark.ml.evaluation import MulticlassClassificationEvaluator
def transData(data):
return data.rdd.map(lambda r: [Vectors.dense(r[:-1]),r[-1]]).toDF(['features','label'])
transformed = transData(df)
transformed.show(5)
+--------------------+-----+
| features|label|
+--------------------+-----+
|[7.4,0.7,0.0,1.9,...| low|
|[7.8,0.88,0.0,2.6...| low|
|[7.8,0.76,0.04,2....| low|
|[11.2,0.28,0.56,1...| low|
|[7.4,0.7,0.0,1.9,...| low|
+--------------------+-----+
only showing top 5 rows
Deal with Categorical Label and Variables
# Index labels, adding metadata to the label column
labelIndexer = StringIndexer(inputCol='label',
outputCol='indexedLabel').fit(transformed)
labelIndexer.transform(transformed).show(5, True)
+--------------------+-----+------------+
| features|label|indexedLabel|
+--------------------+-----+------------+
|[7.4,0.7,0.0,1.9,...| low| 0.0|
|[7.8,0.88,0.0,2.6...| low| 0.0|
|[7.8,0.76,0.04,2....| low| 0.0|
|[11.2,0.28,0.56,1...| low| 0.0|
|[7.4,0.7,0.0,1.9,...| low| 0.0|
+--------------------+-----+------------+
only showing top 5 rows
# Automatically identify categorical features, and index them.
# Set maxCategories so features with > 4 distinct values are treated as continuous.
featureIndexer =VectorIndexer(inputCol="features", \
outputCol="indexedFeatures", \
maxCategories=4).fit(transformed)
featureIndexer.transform(transformed).show(5, True)
+--------------------+-----+--------------------+
| features|label| indexedFeatures|
+--------------------+-----+--------------------+
|[7.4,0.7,0.0,1.9,...| low|[7.4,0.7,0.0,1.9,...|
|[7.8,0.88,0.0,2.6...| low|[7.8,0.88,0.0,2.6...|
|[7.8,0.76,0.04,2....| low|[7.8,0.76,0.04,2....|
|[11.2,0.28,0.56,1...| low|[11.2,0.28,0.56,1...|
|[7.4,0.7,0.0,1.9,...| low|[7.4,0.7,0.0,1.9,...|
+--------------------+-----+--------------------+
only showing top 5 rows
Split the data to training and test data sets
# Split the data into training and test sets (40% held out for testing)
(trainingData, testData) = data.randomSplit([0.6, 0.4])
trainingData.show(5,False)
testData.show(5,False)
+---------------------------------------------------------+-----+
|features |label|
+---------------------------------------------------------+-----+
|[5.0,0.38,0.01,1.6,0.048,26.0,60.0,0.99084,3.7,0.75,14.0]|low |
|[5.0,0.42,0.24,2.0,0.06,19.0,50.0,0.9917,3.72,0.74,14.0] |high |
|[5.0,0.74,0.0,1.2,0.041,16.0,46.0,0.99258,4.01,0.59,12.5]|low |
|[5.0,1.02,0.04,1.4,0.045,41.0,85.0,0.9938,3.75,0.48,10.5]|low |
|[5.0,1.04,0.24,1.6,0.05,32.0,96.0,0.9934,3.74,0.62,11.5] |low |
+---------------------------------------------------------+-----+
only showing top 5 rows
+---------------------------------------------------------+-----+
|features |label|
+---------------------------------------------------------+-----+
|[4.6,0.52,0.15,2.1,0.054,8.0,65.0,0.9934,3.9,0.56,13.1] |low |
|[4.7,0.6,0.17,2.3,0.058,17.0,106.0,0.9932,3.85,0.6,12.9] |low |
|[4.9,0.42,0.0,2.1,0.048,16.0,42.0,0.99154,3.71,0.74,14.0]|high |
|[5.0,0.4,0.5,4.3,0.046,29.0,80.0,0.9902,3.49,0.66,13.6] |low |
|[5.2,0.49,0.26,2.3,0.09,23.0,74.0,0.9953,3.71,0.62,12.2] |low |
+---------------------------------------------------------+-----+
only showing top 5 rows
Fit Naive Bayes Classification Model
from pyspark.ml.classification import NaiveBayes
nb = NaiveBayes(featuresCol='indexedFeatures', labelCol='indexedLabel')
Pipeline Architecture
# Convert indexed labels back to original labels.
labelConverter = IndexToString(inputCol="prediction", outputCol="predictedLabel",
labels=labelIndexer.labels)
# Chain indexers and tree in a Pipeline
pipeline = Pipeline(stages=[labelIndexer, featureIndexer, nb,labelConverter])
# Train model. This also runs the indexers.
model = pipeline.fit(trainingData)
Make predictions
# Make predictions.
predictions = model.transform(testData)
# Select example rows to display.
predictions.select("features","label","predictedLabel").show(5)
+--------------------+-----+--------------+
| features|label|predictedLabel|
+--------------------+-----+--------------+
|[4.6,0.52,0.15,2....| low| low|
|[4.7,0.6,0.17,2.3...| low| low|
|[4.9,0.42,0.0,2.1...| high| low|
|[5.0,0.4,0.5,4.3,...| low| low|
|[5.2,0.49,0.26,2....| low| low|
+--------------------+-----+--------------+
only showing top 5 rows
Evaluation
from pyspark.ml.evaluation import MulticlassClassificationEvaluator
# Select (prediction, true label) and compute test error
evaluator = MulticlassClassificationEvaluator(
labelCol="indexedLabel", predictionCol="prediction", metricName="accuracy")
accuracy = evaluator.evaluate(predictions)
print("Test Error = %g" % (1.0 - accuracy))
Test Error = 0.307339
lrModel = model.stages[2]
trainingSummary = lrModel.summary
# Obtain the objective per iteration
# objectiveHistory = trainingSummary.objectiveHistory
# print("objectiveHistory:")
# for objective in objectiveHistory:
# print(objective)
# Obtain the receiver-operating characteristic as a dataframe and areaUnderROC.
trainingSummary.roc.show(5)
print("areaUnderROC: " + str(trainingSummary.areaUnderROC))
# Set the model threshold to maximize F-Measure
fMeasure = trainingSummary.fMeasureByThreshold
maxFMeasure = fMeasure.groupBy().max('F-Measure').select('max(F-Measure)').head(5)
# bestThreshold = fMeasure.where(fMeasure['F-Measure'] == maxFMeasure['max(F-Measure)']) \
# .select('threshold').head()['threshold']
# lr.setThreshold(bestThreshold)
You can use z.show() to get the data and plot the ROC curves:

You can also register a TempTable data.registerTempTable('roc_data') and then
use sql to plot the ROC curve:

visualization
import matplotlib.pyplot as plt
import numpy as np
import itertools
def plot_confusion_matrix(cm, classes,
normalize=False,
title='Confusion matrix',
cmap=plt.cm.Blues):
"""
This function prints and plots the confusion matrix.
Normalization can be applied by setting `normalize=True`.
"""
if normalize:
cm = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]
print("Normalized confusion matrix")
else:
print('Confusion matrix, without normalization')
print(cm)
plt.imshow(cm, interpolation='nearest', cmap=cmap)
plt.title(title)
plt.colorbar()
tick_marks = np.arange(len(classes))
plt.xticks(tick_marks, classes, rotation=45)
plt.yticks(tick_marks, classes)
fmt = '.2f' if normalize else 'd'
thresh = cm.max() / 2.
for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):
plt.text(j, i, format(cm[i, j], fmt),
horizontalalignment="center",
color="white" if cm[i, j] > thresh else "black")
plt.tight_layout()
plt.ylabel('True label')
plt.xlabel('Predicted label')
class_temp = predictions.select("label").groupBy("label")\
.count().sort('count', ascending=False).toPandas()
class_temp = class_temp["label"].values.tolist()
class_names = map(str, class_temp)
# # # print(class_name)
class_names
['low', 'high']
from sklearn.metrics import confusion_matrix
y_true = predictions.select("label")
y_true = y_true.toPandas()
y_pred = predictions.select("predictedLabel")
y_pred = y_pred.toPandas()
cnf_matrix = confusion_matrix(y_true, y_pred,labels=class_names)
cnf_matrix
array([[392, 169],
[ 32, 61]])
# Plot non-normalized confusion matrix
plt.figure()
plot_confusion_matrix(cnf_matrix, classes=class_names,
title='Confusion matrix, without normalization')
plt.show()
Confusion matrix, without normalization
[[392 169]
[ 32 61]]
# Plot normalized confusion matrix
plt.figure()
plot_confusion_matrix(cnf_matrix, classes=class_names, normalize=True,
title='Normalized confusion matrix')
plt.show()
Normalized confusion matrix
[[0.69875223 0.30124777]
[0.34408602 0.65591398]]